2XX9
 
 | | Crystal structure of 1-((2-fluoro-4-(3-(trifluoromethyl)-4,5,6,7- tetrahydro-1H-indazol-1-yl)phenyl)methyl)-2-pyrrolidinone in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N,N-DIMETHYL-4-[3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOL-1-YL]BENZAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
3IQL
 
 | | Crystal structure of the rat endophilin-A1 SH3 domain | | Descriptor: | CHLORIDE ION, Endophilin-A1 | | Authors: | Trempe, J.F, Kozlov, G, Camacho, E.M, Gehring, K. | | Deposit date: | 2009-08-20 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | SH3 domains from a subset of BAR proteins define a Ubl-binding domain and implicate parkin in synaptic ubiquitination.
Mol.Cell, 36, 2009
|
|
8I40
 
 | | Crystal structure of ASCT from Trypanosoma brucei in complex with CoA. | | Descriptor: | ACETATE ION, CALCIUM ION, COENZYME A, ... | | Authors: | Mochizuki, K, Inaoka, D.K, Fukuda, K, Kurasawa, H, Iyoda, K, Nakai, U, Harada, S, Balogun, E.O, Mazet, M, Millerioux, Y, Bringaud, F, Boshart, M, Hirayama, K, Kita, K, Shiba, T. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of ligand complexes of ASCT from Trypanosoma brucei and molecular mechanism in comparison with mammalian SCOT.
To Be Published
|
|
8RWL
 
 | | Crystal structure of Methanopyrus kandleri malate dehydrogenase mutant 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Malate dehydrogenase, ... | | Authors: | Coquille, S, Roche, J, Engilberge, S, Girard, E, Madern, D. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Navigating the conformational landscape of an enzyme. Stabilization of a low populated conformer by evolutionary mutations triggers Allostery into a non-allosteric enzyme.
To Be Published
|
|
7EXD
 
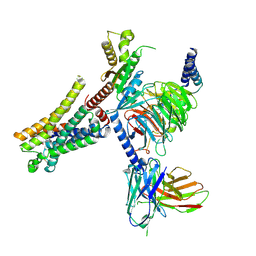 | | Lasmiditan-bound serotonin 1F (5-HT1F) receptor-Gi protein complex | | Descriptor: | 2,4,6-tris(fluoranyl)-N-[6-(1-methylpiperidin-4-yl)carbonylpyridin-2-yl]benzamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S, Xu, P, Jiang, Y, Xu, H.E. | | Deposit date: | 2021-05-27 | | Release date: | 2021-08-04 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for recognition of anti-migraine drug lasmiditan by the serotonin receptor 5-HT 1F -G protein complex.
Cell Res., 31, 2021
|
|
5L5F
 
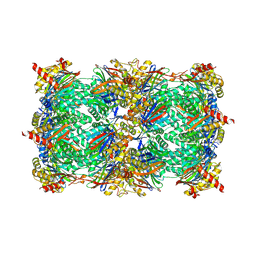 | |
1OHF
 
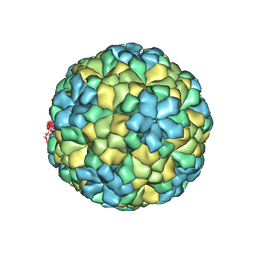 | | The refined structure of Nudaurelia capensis omega virus | | Descriptor: | MAGNESIUM ION, p70 | | Authors: | Helgstrand, C, Munshi, S, Johnson, J.E, Liljas, L. | | Deposit date: | 2003-05-26 | | Release date: | 2004-02-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Refined Structure of Nudaurelia Capensis Omega Virus Reveals Control Elements for a T = 4 Capsid Maturation
Virology, 318, 2004
|
|
6MA4
 
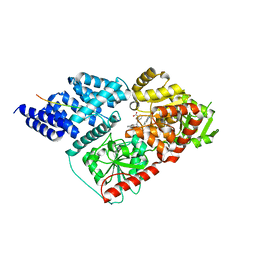 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 3a | | Descriptor: | 5-{2-[(1R)-2-{(carboxymethyl)[(thiophen-2-yl)methyl]amino}-2-oxo-1-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}ethyl]phenoxy}pentanoic acid, Host Cell Factor 1 peptide, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
5L5Z
 
 | |
5L54
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 16 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
5L6C
 
 | |
2UXW
 
 | | Crystal structure of human very long chain acyl-CoA dehydrogenase (ACADVL) | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, TRANS DELTA2 PALMITENOYL-COENZYMEA, ... | | Authors: | Pike, A.C.W, Hozjan, V, Smee, C, Berridge, G, Burgess, N, Salah, E, Bunkoczi, G, Uppenberg, J, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Oppermann, U. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Human Very Long Chain Acyl- Coa Dehydrogenase (Acadvl)
To be Published
|
|
5L5T
 
 | | Yeast 20S proteasome with human beta5i (1-138; V31M) and human beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 16 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
6SPP
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase variant VI298 | | Descriptor: | CITRIC ACID, GLYCEROL, Methionine--tRNA ligase, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
5L68
 
 | |
6MBD
 
 | | Human Mcl-1 in complex with the designed peptide dM1 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION, dM1 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
5L8F
 
 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E32A, E62A, H65A. | | Descriptor: | MAGNESIUM ION, Rru_A0973, TRIETHYLENE GLYCOL | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E32A, E62A, H65A.
To be published
|
|
6MBO
 
 | | GLP Methyltransferase with Inhibitor EML741-P212121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-7-methoxy-N-[1-(propan-2-yl)piperidin-4-yl]-8-[3-(pyrrolidin-1-yl)propoxy]-3H-1,4-benzodiazepin-5-amine, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Discovery of a Novel Chemotype of Histone Lysine Methyltransferase EHMT1/2 (GLP/G9a) Inhibitors: Rational Design, Synthesis, Biological Evaluation, and Co-crystal Structure.
J. Med. Chem., 62, 2019
|
|
6MGB
 
 | | Thermosulfurimonas dismutans KpsC, beta Kdo 2,4 transferase | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, Capsular polysaccharide export system protein KpsC, ... | | Authors: | Doyle, L, Mallette, E, Kimber, M.S, Whitfield, C. | | Deposit date: | 2018-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biosynthesis of a conserved glycolipid anchor for Gram-negative bacterial capsules.
Nat.Chem.Biol., 15, 2019
|
|
6MA1
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 4a | | Descriptor: | Host Cell Factor 1 peptide, N-[(2R)-2-{[(7-chloro-2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}-2-(2-methoxyphenyl)acetyl]-N-[(thiophen-2-yl)methyl]glycine, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
7EZM
 
 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gq complex | | Descriptor: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
7EZH
 
 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gi complex | | Descriptor: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
5EYV
 
 | | Crystal Structure of Adenylosuccinate lyase from Schistosoma mansoni in APO form. | | Descriptor: | Adenylosuccinate lyase | | Authors: | Romanello, L, Torini, J.R, Bird, L.E, Nettleship, J.E, Owens, R.J, Reddivari, Y, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural and kinetic analysis of Schistosoma mansoni Adenylosuccinate Lyase (SmADSL).
Mol. Biochem. Parasitol., 214, 2017
|
|
8SZE
 
 | | Crystal structure of Yersinia pestis dihydrofolate reductase in complex with Trimethoprim | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Shaw, G.X, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2023-05-29 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Yersinia pestis dihydrofolate reductase in complex with Trimethoprim
To be published
|
|
8TLI
 
 | | THERMOLYSIN (100% ISOPROPANOL SOAKED CRYSTALS) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1998-10-30 | | Release date: | 2000-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
