4X7G
 
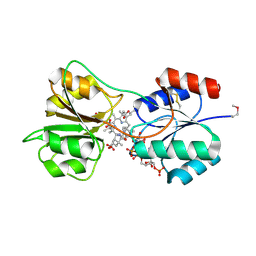 | | CobK precorrin-6A reductase | | Descriptor: | 3-[(1R,2S,3S,7S,11S,16S,17R,18R,19R)-2,7,12,18-tetrakis(2-hydroxy-2-oxoethyl)-3,13-bis(3-hydroxy-3-oxopropyl)-1,2,5,7,11,17-hexamethyl-17-(3-oxidanyl-3-oxidanylidene-prop-1-enyl)-3,6,8,10,15,16,18,19,21,24-decahydrocorrin-8-yl]propanoic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Precorrin-6A reductase | | Authors: | Gu, S, Deery, E, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2014-12-09 | | Release date: | 2016-01-20 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal structure of CobK reveals strand-swapping between Rossmann-fold domains and molecular basis of the reduced precorrin product trap.
Sci Rep, 5, 2015
|
|
8S9T
 
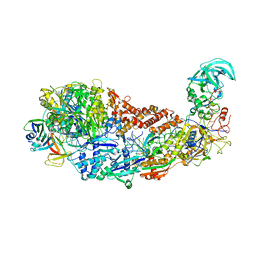 | | CRISPR-Cas type III-D effector complex | | Descriptor: | CRISPR RNA, Cas10, Cas7-2x, ... | | Authors: | Schwartz, E.A, Taylor, D.W. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | RNA targeting and cleavage by the type III-Dv CRISPR effector complex.
Nat Commun, 15, 2024
|
|
4LOG
 
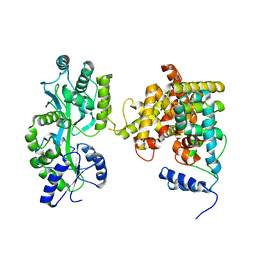 | | The crystal structure of the orphan nuclear receptor PNR ligand binding domain fused with MBP | | Descriptor: | Maltose ABC transporter periplasmic protein and NR2E3 protein chimeric construct | | Authors: | Tan, M.E, Zhou, X.E, Soon, F.-F, Li, X, Li, J, Yong, E.-L, Melcher, K, Xu, H.E. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of the Orphan Nuclear Receptor NR2E3/PNR Ligand Binding Domain Reveals a Dimeric Auto-Repressed Conformation.
Plos One, 8, 2013
|
|
6S70
 
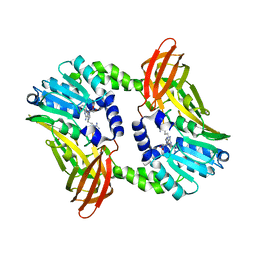 | | Crystal structure of CARM1 in complex with inhibitor UM251 | | Descriptor: | 1-[5-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]pentyl]guanidine, GLYCEROL, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
8S9V
 
 | |
6S79
 
 | | Crystal structure of CARM1 in complex with inhibitor AA183 | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-(pyridin-2-ylamino)propyl]amino]-2-azanyl-butanoic acid, GLYCEROL, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Al-Noori, A, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
6S2B
 
 | | Structure of beta-fructofuranosidase from Schwanniomyces occidentalis complexed with fructosyl-erythritol | | Descriptor: | (2~{S},3~{R})-4-[(2~{R},3~{S},4~{S},5~{R})-2,5-bis(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxybutane-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2019-06-20 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | New insights into the molecular mechanism behind mannitol and erythritol fructosylation by beta-fructofuranosidase from Schwanniomyces occidentalis.
Sci Rep, 11, 2021
|
|
8S9X
 
 | |
6S86
 
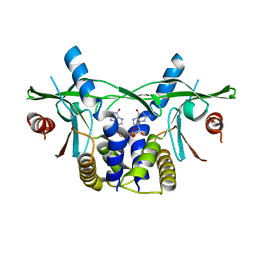 | | Human wtSTING in complex with 3'3'-c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Stimulator of Interferon genes | | Authors: | Smola, M, Boura, E. | | Deposit date: | 2019-07-08 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | No magnesium is needed for binding of the stimulator of interferon genes to cyclic dinucleotides.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4WJL
 
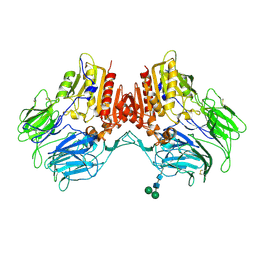 | | Structure of human dipeptidyl peptidase 10 (DPPY): a modulator of neuronal Kv4 channels | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Inactive dipeptidyl peptidase 10, ... | | Authors: | Bezerra, G.A, Dobrovetsky, E, Seitova, A, Fedosyuk, S, Dhe-Paganon, S, Gruber, K. | | Deposit date: | 2014-09-30 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of human dipeptidyl peptidase 10 (DPPY): a modulator of neuronal Kv4 channels.
Sci Rep, 5, 2015
|
|
6MAX
 
 | |
5L5H
 
 | |
7KTX
 
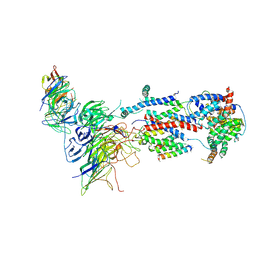 | | Cryo-EM structure of Saccharomyces cerevisiae ER membrane protein complex bound to a Fab in DDM detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 2, ... | | Authors: | Miller-Vedam, L.E, Schirle Oakdale, N.S, Braeuning, B, Boydston, E.A, Sevillano, N, Popova, K.D, Bonnar, J.L, Shurtleff, M.J, Prabu, J.R, Stroud, R.M, Craik, C.S, Schulman, B.A, Weissman, J.S, Frost, A. | | Deposit date: | 2020-11-24 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
5L5V
 
 | |
8SCA
 
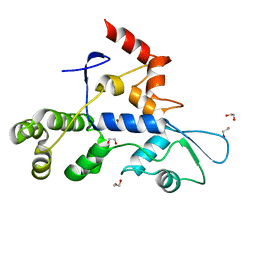 | | Rec3 Domain from S. pyogenes Cas9 | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1 | | Authors: | D'Ordine, A.M, Skeens, E, Lisi, G.P, Jogl, G. | | Deposit date: | 2023-04-05 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | High-fidelity, hyper-accurate, and evolved mutants rewire atomic-level communication in CRISPR-Cas9.
Sci Adv, 10, 2024
|
|
5L6B
 
 | | Yeast 20S proteasome with mouse beta5i (1-138) and mouse beta6 (97-111; 118-133) in complex with ONX 0914 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
4HIB
 
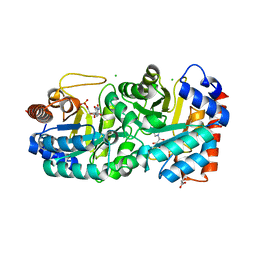 | | Crystal structure of human orotidine 5'-monophosphate decarboxylase complexed with CMP-N4-OH | | Descriptor: | CHLORIDE ION, GLYCEROL, N-hydroxycytidine 5'-(dihydrogen phosphate), ... | | Authors: | To, T.K, Kotra, L.P, Pai, E.F. | | Deposit date: | 2012-10-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel cytidine-based orotidine-5'-monophosphate decarboxylase inhibitors with an unusual twist.
J.Med.Chem., 55, 2012
|
|
8SAG
 
 | |
1JA8
 
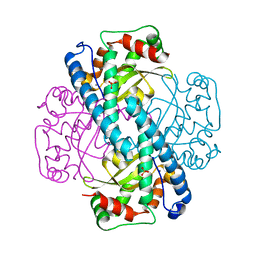 | | Kinetic Analysis of Product Inhibition in Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Manganese Superoxide Dismutase, SULFATE ION | | Authors: | Hearn, A.S, Stroupe, M.E, Cabelli, D.E, Lepock, J.R, Tainer, J.A, Nick, H.S, Silverman, D.S. | | Deposit date: | 2001-05-29 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Kinetic analysis of product inhibition in human manganese superoxide dismutase.
Biochemistry, 40, 2001
|
|
1JC4
 
 | | Crystal Structure of Se-Met Methylmalonyl-CoA Epimerase | | Descriptor: | Methylmalonyl-CoA epimerase, SULFATE ION | | Authors: | Mc Carthy, A.A, Baker, H.M, Shewry, S.C, Patchett, M.L, Baker, E.N. | | Deposit date: | 2001-06-07 | | Release date: | 2001-07-11 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of methylmalonyl-coenzyme A epimerase from P. shermanii: a novel enzymatic function on an ancient metal binding scaffold.
Structure, 9, 2001
|
|
8SBF
 
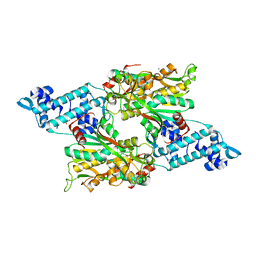 | | Full-length structure of the LysR-type transcriptional regulator, ACIAD0746, from Acinetobacter baylyi | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative transcriptional regulator (LysR family), ... | | Authors: | Momany, C, Nune, M, Brondani, J.C, Afful, D, Neidle, E, Galloway, N.R. | | Deposit date: | 2023-04-03 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | FinR, a LysR-type transcriptional regulator involved in sulfur homeostasis with homologs in diverse microorganisms
To Be Published
|
|
6LPJ
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 277 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
7Q49
 
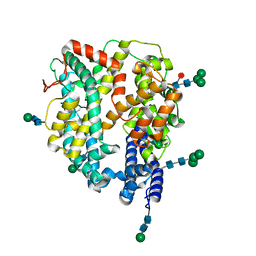 | | Local refinement structure of the N-domain of full-length, monomeric, soluble somatic angiotensin I-converting enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ZINC ION, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-29 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
7Q3Y
 
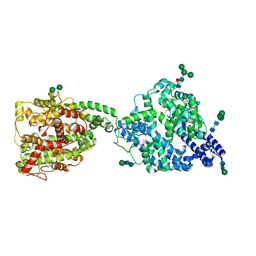 | | Structure of full-length, monomeric, soluble somatic angiotensin I-converting enzyme showing the N- and C-terminal ellipsoid domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-29 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
7VFP
 
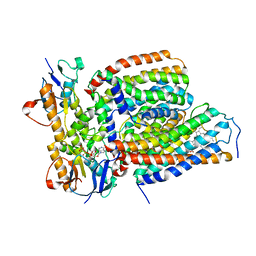 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with heme and single ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-09-13 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
