8FSQ
 
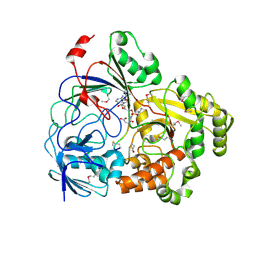 | | Complex Structure of YejA with Microcin C7 | | Descriptor: | 5'-O-[(R)-amino(3-aminopropoxy)phosphoryl]adenosine, Microcin C7 peptide portion, YejA | | Authors: | Naik, S.K, Dong, S.-H. | | Deposit date: | 2023-01-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Trojan Horse Peptide Conjugates Remodel the Activity Spectrum of Clinical Antibiotics
To Be Published
|
|
8P2K
 
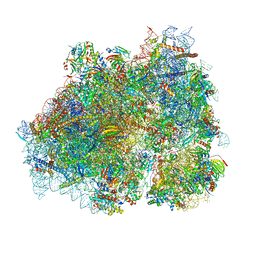 | | Ternary complex of translating ribosome, NAC and METAP1 | | Descriptor: | 18s rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Jia, M, Jaskolowski, M, Scaiola, A, Jomaa, A, Ban, N. | | Deposit date: | 2023-05-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | NAC controls cotranslational N-terminal methionine excision in eukaryotes.
Science, 380, 2023
|
|
5NIT
 
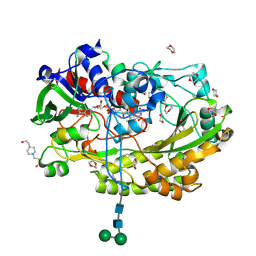 | | Glucose oxidase mutant A2 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Hoffmann, K. | | Deposit date: | 2017-03-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Shuffling Active Site Substate Populations Affects Catalytic Activity: The Case of Glucose Oxidase.
ACS Catal, 7, 2017
|
|
7JRS
 
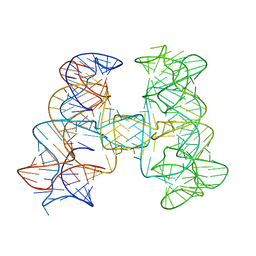 | |
1FWU
 
 | |
1W0G
 
 | | Crystal structure of human cytochrome P450 3A4 | | Descriptor: | CYTOCHROME P450 3A4, METYRAPONE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Williams, P.A, Cosme, J, Vinkovic, D.M, Ward, A, Angove, H.C, Day, P.J, Vonrhein, C, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-06-03 | | Release date: | 2004-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structures of Human Cytochrome P450 3A4 Bound to Metyrapone and Progesterone
Science, 305, 2004
|
|
5YPS
 
 | | The structural basis of histone chaperoneVps75 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | Deposit date: | 2017-11-03 | | Release date: | 2018-11-07 | | Last modified: | 2020-06-10 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
7KC0
 
 | | Structure of the Saccharomyces cerevisiae replicative polymerase delta in complex with a primer/template and the PCNA clamp | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA (25-MER), DNA (5'-D(P*AP*TP*GP*AP*CP*CP*AP*TP*GP*AP*TP*TP*AP*CP*GP*AP*AP*TP*TP*GP*C)-3'), ... | | Authors: | Zheng, F, Georgescu, R, Li, H, O'Donnell, M.E. | | Deposit date: | 2020-10-04 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of eukaryotic DNA polymerase delta bound to the PCNA clamp while encircling DNA.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YIC
 
 | |
6V35
 
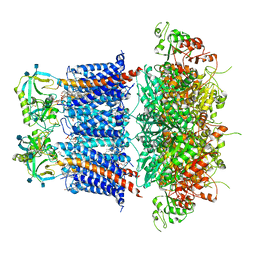 | | Cryo-EM structure of Ca2+-free hsSlo1-beta4 channel complex | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Tao, X, MacKinnon, R. | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular structures of the human Slo1 K + channel in complex with beta 4.
Elife, 8, 2019
|
|
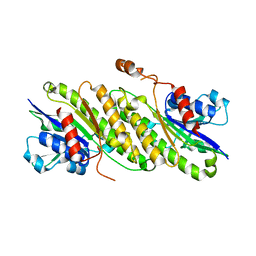
4FD3
 
 | |
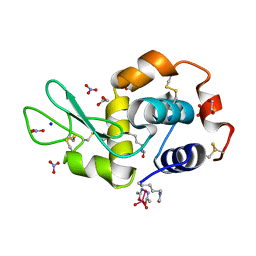
5NJ1
 
 | |
4L9K
 
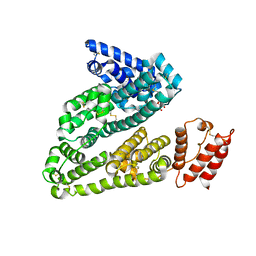 | | X-ray study of human serum albumin complexed with camptothecin | | Descriptor: | (2S)-2-hydroxy-2-[8-(hydroxymethyl)-9-oxo-9,11-dihydroindolizino[1,2-b]quinolin-7-yl]butanoic acid, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-18 | | Release date: | 2013-07-24 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
6YIZ
 
 | |
5ABU
 
 | |
6YRV
 
 | | Crystal structure of FAP after illumination at 100K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
7JPS
 
 | |
6YRU
 
 | | Crystal structure of FAP in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
5AAG
 
 | | Aurora A kinase bound to an imidazopyridine inhibitor (14b) | | Descriptor: | AURORA KINASE A, [3-[[4-[6-chloranyl-2-(1,3-dimethylpyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl]pyrazol-1-yl]methyl]phenyl]-(4-methylpiperazin-1-yl)methanone | | Authors: | McIntyre, P.J, Bayliss, R. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 7-(Pyrazol-4-Yl)-3H-Imidazo[4,5-B]Pyridine-Based Derivatives for Kinase Inhibition: Co-Crystallisation Studies with Aurora-A Reveal Distinct Differences in the Orientation of the Pyrazole N1-Substituent.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5M10
 
 | |
6V4S
 
 | |
8GRV
 
 | |
5M16
 
 | | Structure of GH36 alpha-galactosidase from Thermotoga maritima in complex with a hydrolysed cyclopropyl carbasugar. | | Descriptor: | (1~{R},2~{S},3~{S},4~{S},5~{S},6~{S})-1-(hydroxymethyl)bicyclo[4.1.0]heptane-2,3,4,5-tetrol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Pengelly, R, Gloster, T. | | Deposit date: | 2016-10-07 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Snapshots for Mechanism-Based Inactivation of a Glycoside Hydrolase by Cyclopropyl Carbasugars.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
1FYT
 
 | | CRYSTAL STRUCTURE OF A COMPLEX OF A HUMAN ALPHA/BETA-T CELL RECEPTOR, INFLUENZA HA ANTIGEN PEPTIDE, AND MHC CLASS II MOLECULE, HLA-DR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ HA1 PEPTIDE CHAIN, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Hennecke, J, Carfi, A, Wiley, D.C. | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a covalently stabilized complex of a human alphabeta T-cell receptor, influenza HA peptide and MHC class II molecule, HLA-DR1.
EMBO J., 19, 2000
|
|
6YON
 
 | | Crystal structure of Endoglucanase S127C/A165C from Penicillium verruculosum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoglucanase, SULFATE ION | | Authors: | Nemashkalov, V, Kravchenko, O, Gabdulkhakov, A, Tischenko, S, Rozhkova, A, Sinitsyn, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Endoglucanase S127C/A165C from Penicillium verruculosum
To Be Published
|
|
