5JIB
 
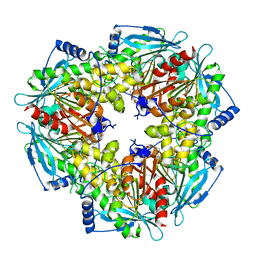 | |
7V2L
 
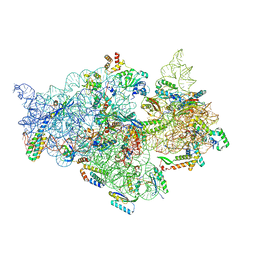 | | T.thermophilus 30S ribosome with KsgA, class K1k2 | | Descriptor: | 16s ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Raina, R, Singh, J, Anand, R, Vinothkumar, K.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
7V2P
 
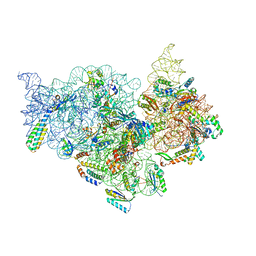 | | T.thermophilus 30S ribosome with KsgA, class K5 | | Descriptor: | 16s ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Raina, R, Singh, J, Anand, R, Vinothkumar, K.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Decoding the Mechanism of Specific RNA Targeting by Ribosomal Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
8RF2
 
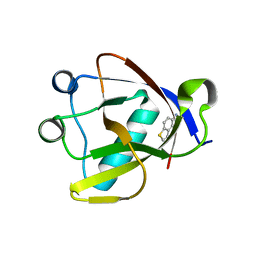 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 1E7 refined against the anomalous diffraction data | | Descriptor: | 1-benzothiophen-5-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7K95
 
 | | Crystal structure of human CPSF30 in complex with hFip1 | | Descriptor: | Isoform 2 of Cleavage and polyadenylation specificity factor subunit 4, Pre-mRNA 3'-end-processing factor FIP1, ZINC ION | | Authors: | Hamilton, K, Tong, L. | | Deposit date: | 2020-09-28 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism for the interaction between human CPSF30 and hFip1.
Genes Dev., 34, 2020
|
|
7K4Z
 
 | | Crystal structure of Kemp Eliminase HG3.17 in complex with the transition state analog 6-nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, PENTAETHYLENE GLYCOL | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
5JJL
 
 | | Rho transcription termination factor bound to rU8 and 5 ADP-BeF3 molecules | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Thomsen, N.D, Lawson, M.R, Witkowsky, L.B, Qu, S, Berger, J.M. | | Deposit date: | 2016-04-24 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular mechanisms of substrate-controlled ring dynamics and substepping in a nucleic acid-dependent hexameric motor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
3MP2
 
 | |
8RFC
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2_AL1 refined against the anomalous diffraction data | | Descriptor: | (1~{R})-1-(4-bromophenyl)ethanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RGL
 
 | | Structure of Human Serum Albumin in complex with Aristolochic Acid I at 1.9 A resolution - Optimized | | Descriptor: | 1,2-ETHANEDIOL, 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, MYRISTIC ACID, ... | | Authors: | Pomyalov, S, Sidorenko, V.S, Grollman, A.P, Shoham, G. | | Deposit date: | 2023-12-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic insights into the transport of aristolochic acids and their active metabolites by human serum albumin.
J.Biol.Chem., 300, 2024
|
|
5JIG
 
 | | Crytsal structure of Wss1 from S. pombe | | Descriptor: | NICKEL (II) ION, OXYGEN MOLECULE, Ubiquitin and WLM domain-containing metalloprotease SPCC1442.07c | | Authors: | Groll, M, Stingele, J, Boulton, S. | | Deposit date: | 2016-04-22 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Mechanism and Regulation of DNA-Protein Crosslink Repair by the DNA-Dependent Metalloprotease SPRTN.
Mol.Cell, 64, 2016
|
|
3MSD
 
 | | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. | | Descriptor: | ACETATE ION, GLYCEROL, Intra-cellular xylanase ixt6, ... | | Authors: | Solomon, V, Zolotnitsky, G, Alhadeff, R, Shoham, Y, Shoham, G. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus.
TO BE PUBLISHED
|
|
8RFD
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2_AL2 refined against the anomalous diffraction data | | Descriptor: | (1~{R})-1-(4-iodophenyl)ethanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
3M0M
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329F in complex with D-allose | | Descriptor: | D-ALLOSE, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
8RGK
 
 | | Structure of Human Serum Albumin in complex with Aristolochic Acid at 1.9 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, MYRISTIC ACID, ... | | Authors: | Pomyalov, S, Sidorenko, V.S, Grollman, A.P, Shoham, G. | | Deposit date: | 2023-12-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic insights into the transport of aristolochic acids and their active metabolites by human serum albumin.
J.Biol.Chem., 300, 2024
|
|
8RF6
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11A7_AL5 refined against the anomalous diffraction data | | Descriptor: | 6-iodanyl-2,3-dihydro-1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
4ZJL
 
 | |
1Z7C
 
 | |
7JPV
 
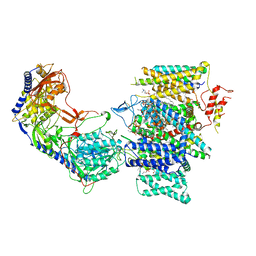 | | Rabbit Cav1.1 in the presence of 1 micromolar (S)-(-)-Bay K8644 in nanodiscs at 3.4 Angstrom resolution | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, CALCIUM ION, ... | | Authors: | Yan, N, Gao, S. | | Deposit date: | 2020-08-10 | | Release date: | 2020-11-18 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis of the Modulation of the Voltage-Gated Calcium Ion Channel Ca v 1.1 by Dihydropyridine Compounds*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7UNN
 
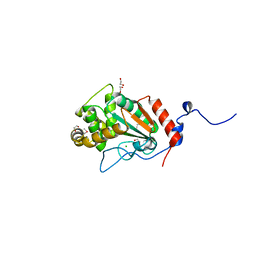 | | Thiol-disulfide oxidoreductase TsdA from Corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Thioredoxin domain-containing protein | | Authors: | Osipiuk, J, Reardon-Robinson, M, Nguyen, M.T, Sanchez, B, Ton-That, H, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-11 | | Release date: | 2022-04-20 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A cryptic oxidoreductase safeguards oxidative protein folding in Corynebacterium diphtheriae.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8RF5
 
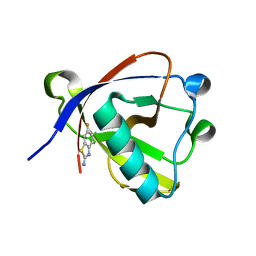 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11A7 refined against the anomalous diffraction data | | Descriptor: | 6-fluoro-1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7JRG
 
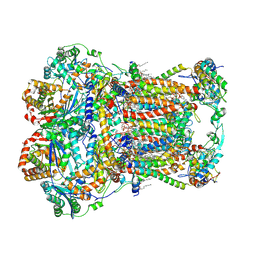 | | Plant Mitochondrial complex III2 from Vigna radiata | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Alpha-MPP, ... | | Authors: | Maldonado, M, Letts, J.A. | | Deposit date: | 2020-08-12 | | Release date: | 2021-01-20 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Atomic structures of respiratory complex III 2 , complex IV, and supercomplex III 2 -IV from vascular plants.
Elife, 10, 2021
|
|
4ZLN
 
 | |
7JX9
 
 | | The crystal structure of human ornithine aminotransferase with an intermediate bound during inactivation by (1S,3S)-3-amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic acid. | | Descriptor: | (1S,3S,4S)-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-(1,1,3,3,3-pentafluoroprop-1-en-2-yl)cyclopentane-1-carboxylic acid, N-[1,3-dihydroxy-2-(hydroxymethyl)propan-2-yl]glycine, Ornithine aminotransferase, ... | | Authors: | Butrin, A, Beaupre, B, Shen, S, Silverman, R.B, Moran, G, Liu, D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Kinetic Analyses Reveal the Dual Inhibition Modes of Ornithine Aminotransferase by (1 S ,3 S )-3-Amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic Acid (BCF 3 ).
Acs Chem.Biol., 16, 2021
|
|
8RF3
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7G3 refined against the anomalous diffraction data | | Descriptor: | 2-(1-benzothiophen-3-yl)ethanoic acid, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
