5TJS
 
 | |
5TK3
 
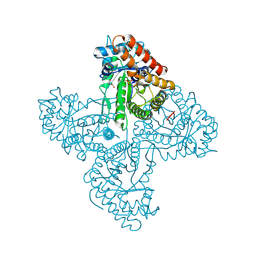 | | Crystal structure of FBP aldolase from Toxoplasma gondii, burst-phase ternary complex | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Heron, P.W, Sygusch, J. | | Deposit date: | 2016-10-06 | | Release date: | 2017-10-04 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Isomer activation controls stereospecificity of class I fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 292, 2017
|
|
5TKN
 
 | |
6CNA
 
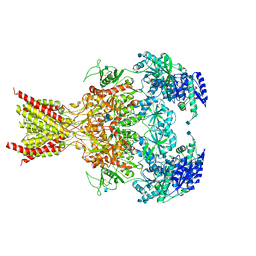 | | GluN1-GluN2B NMDA receptors with exon 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Furukawa, H, Grant, T, Grigorieff, N. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Mechanism of Functional Modulation by Gene Splicing in NMDA Receptors.
Neuron, 98, 2018
|
|
3ZFH
 
 | | Crystal structure of Pseudomonas aeruginosa inosine 5'-monophosphate dehydrogenase | | Descriptor: | CHLORIDE ION, INOSINE 5'-MONOPHOSPHATE DEHYDROGENASE | | Authors: | Rao, V.A, Shepherd, S.M, Owen, R, Hunter, W.N. | | Deposit date: | 2012-12-11 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Pseudomonas Aeruginosa Inosine 5'-Monophosphate Dehydrogenase
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3JUJ
 
 | |
5YZA
 
 | | Crystal Structure of Human CRMP-2 with S522D mutation | | Descriptor: | Dihydropyrimidinase-related protein 2 | | Authors: | Sumi, T, Imasaki, T, Aoki, M, Sakai, N, Nitta, E, Shirouzu, M, Nitta, R. | | Deposit date: | 2017-12-13 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into the Altering Function of CRMP2 by Phosphorylation.
Cell Struct. Funct., 43, 2018
|
|
5CNA
 
 | | REFINED STRUCTURE OF CONCANAVALIN A COMPLEXED WITH ALPHA-METHYL-D-MANNOPYRANOSIDE AT 2.0 ANGSTROMS RESOLUTION AND COMPARISON WITH THE SACCHARIDE-FREE STRUCTURE | | Descriptor: | CALCIUM ION, CHLORIDE ION, CONCANAVALIN A, ... | | Authors: | Naismith, J.H, Emmerich, C, Habash, J, Harrop, S.J, Helliwell, J.R, Hunter, W.N, Raftery, J, Kalb(Gilboa), A.J, Yariv, J. | | Deposit date: | 1994-02-11 | | Release date: | 1994-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structure of concanavalin A complexed with methyl alpha-D-mannopyranoside at 2.0 A resolution and comparison with the saccharide-free structure.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
5TOV
 
 | |
5TKP
 
 | |
6CYJ
 
 | | Mycobacterium tuberculosis transcriptional regulator | | Descriptor: | HTH-type transcriptional regulator PrpR, IRON/SULFUR CLUSTER, SUCCINYL-COENZYME A | | Authors: | Tang, S, Sacchettini, J. | | Deposit date: | 2018-04-05 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structural and functional insight into the Mycobacterium tuberculosis protein PrpR reveals a novel type of transcription factor.
Nucleic Acids Res., 47, 2019
|
|
6CZ6
 
 | | Mycobacterium tuberculosis transcriptional regulator | | Descriptor: | COENZYME A, HTH-type transcriptional regulator PrpR, IRON/SULFUR CLUSTER | | Authors: | Tang, S, Sacchettini, J. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-17 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional insight into the Mycobacterium tuberculosis protein PrpR reveals a novel type of transcription factor.
Nucleic Acids Res., 47, 2019
|
|
3JUK
 
 | |
5D21
 
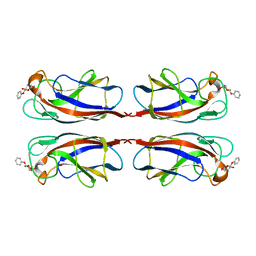 | | Multivalency Effects in Glycopeptide Dendrimer Inhibitors of Pseudomonas aeruginosa Biofilms Targeting Lectin LecA | | Descriptor: | CALCIUM ION, LecA, phenyl beta-D-galactopyranoside | | Authors: | Bergmann, M, Michaud, G, Visini, R, Jin, X, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multivalency effects on Pseudomonas aeruginosa biofilm inhibition and dispersal by glycopeptide dendrimers targeting lectin LecA.
Org.Biomol.Chem., 14, 2016
|
|
5WCH
 
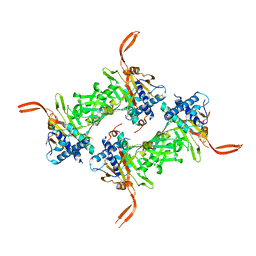 | | Crystal structure of the catalytic domain of human USP9X | | Descriptor: | Probable ubiquitin carboxyl-terminal hydrolase FAF-X, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Zhang, Q, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and activity-based labeling reveal the mechanisms for linkage-specific substrate recognition by deubiquitinase USP9X.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5WYF
 
 | | Structure of amino acid racemase, 2.12 A | | Descriptor: | CADMIUM ION, Isoleucine 2-epimerase, N-[O-PHOSPHONO-PYRIDOXYL]-ISOLEUCINE | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2017-01-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5WQL
 
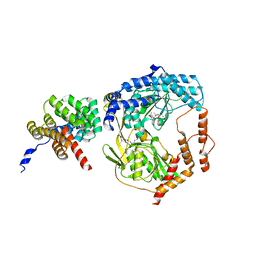 | |
1TRZ
 
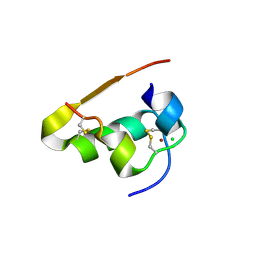 | |
1YFZ
 
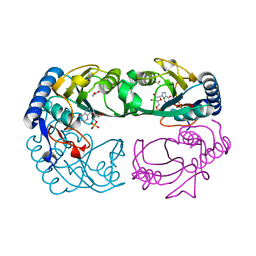 | | Novel IMP Binding in Feedback Inhibition of Hypoxanthine-Guanine Phosphoribosyltransferase from Thermoanaerobacter tengcongensis | | Descriptor: | ACETATE ION, Hypoxanthine-guanine phosphoribosyltransferase, INOSINIC ACID, ... | | Authors: | Chen, Q, Liang, Y, Su, X, Gu, X, Zheng, X, Luo, M. | | Deposit date: | 2005-01-04 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Alternative IMP Binding in Feedback Inhibition of Hypoxanthine-Guanine Phosphoribosyltransferase from Thermoanaerobacter tengcongensis.
J.Mol.Biol., 348, 2005
|
|
1TYM
 
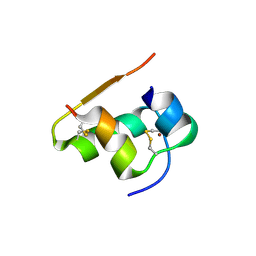 | |
4ADZ
 
 | |
5A6Q
 
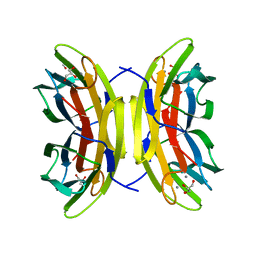 | | Native structure of the LecB lectin from Pseudomonas aeruginosa strain PA14 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, ... | | Authors: | Sommer, R, Wagner, S, Varrot, A, Khaledi, A, Haussler, S, Imberty, A, Titz, A. | | Deposit date: | 2015-06-30 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The virulence factor LecB varies in clinical isolates: consequences for ligand binding and drug discovery.
Chem Sci, 7, 2016
|
|
4AUE
 
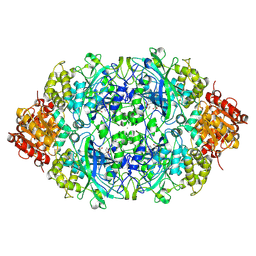 | | Crystal structure, recombinant expression and mutagenesis studies of the bifunctional catalase-phenol oxidase from Scytalidium thermophilum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yuzugullu, Y, Trinh, C.H, Smith, M.A, Pearson, A.R, Phillips, S.E.V, Sutay Kocabas, D, Bakir, U, Ogel, Z.B, McPherson, M.J. | | Deposit date: | 2012-05-17 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure, Recombinant Expression and Mutagenesis Studies of the Catalase with Oxidase Activity from Scytalidium Thermophilum
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5T4D
 
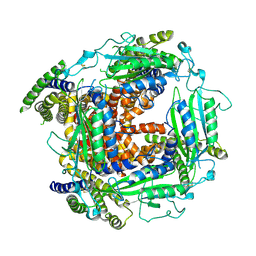 | | Cryo-EM structure of Polycystic Kidney Disease protein 2 (PKD2), residues 198-703 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, hPKD:198-703, Polycystin-2 | | Authors: | Shen, P.S, Yang, X, DeCaen, P.G, Liu, X, Bulkley, D, Clapham, D.E, Cao, E. | | Deposit date: | 2016-08-29 | | Release date: | 2016-11-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Structure of the Polycystic Kidney Disease Channel PKD2 in Lipid Nanodiscs.
Cell, 167, 2016
|
|
7LRD
 
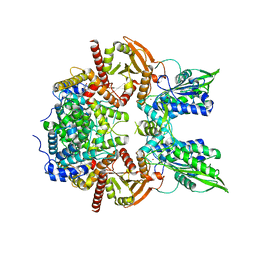 | | Cryo-EM of the SLFN12-PDE3A complex: Consensus subset model | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
