6HW1
 
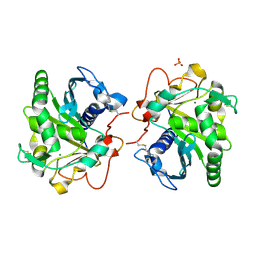 | | ROOM TEMPERATURE STRUCTURE OF LIPASE FROM T. LANUGINOSA AT 2.5 A RESOLUTION IN CHIPX MICROFLUIDIC DEVICE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase, MAGNESIUM ION, ... | | Authors: | Gavira, J.A, Fernadez-Penas, R, Martinez-Rodriguez, S, Verdugo-Escamilla, C. | | Deposit date: | 2018-10-11 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
1PVR
 
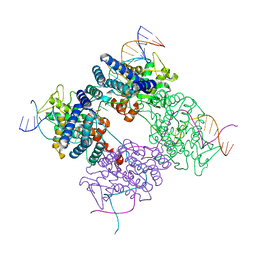 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, LNSGG BOUND TO THE LOXP (WILDTYPE) RECOGNITION SITE | | Descriptor: | 34-MER, Recombinase CRE | | Authors: | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | Deposit date: | 2003-06-28 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
2RAT
 
 | |
1Q5X
 
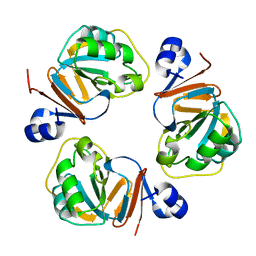 | | Structure of OF RRAA (MENG), a protein inhibitor of RNA processing | | Descriptor: | REGULATOR OF RNASE E ACTIVITY A | | Authors: | Monzingo, A.F, Gao, J, Qiu, J, Georgiou, G, Robertus, J.D. | | Deposit date: | 2003-08-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray Structure of Escherichia coli RraA (MenG), A Protein Inhibitor of RNA Processing.
J.Mol.Biol., 332, 2003
|
|
4JRA
 
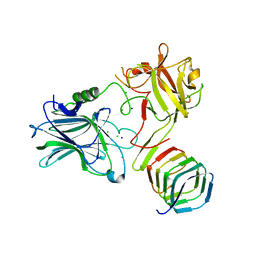 | | CRYSTAL STRUCTURE OF THE BOTULINUM NEUROTOXIN A RECEPTOR-BINDING DOMAIN IN COMPLEX WITH THE LUMINAL DOMAIN Of SV2C | | Descriptor: | Botulinum neurotoxin type A, CHLORIDE ION, SODIUM ION, ... | | Authors: | Benoit, R.M, Frey, D, Wieser, M.M, Jaussi, R, Schertler, G.F.X, Capitani, G, Kammerer, R.A. | | Deposit date: | 2013-03-21 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of synaptic vesicle protein 2C by botulinum neurotoxin A.
Nature, 505, 2014
|
|
1PPJ
 
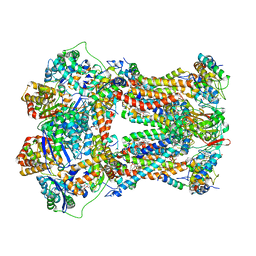 | | Bovine cytochrome bc1 complex with stigmatellin and antimycin | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-METHYL-BUTYRIC ACID 3-(3-FORMYLAMINO-2-HYDROXY-BENZOYLAMINO)-8-HEPTYL-2,6-DIMETHYL-4,9-DIOXO-[1,5]DIOXONAN-7-YL ESTER, AZIDE ION, ... | | Authors: | Huang, L.S, Cobessi, D, Tung, E.Y, Berry, E.A. | | Deposit date: | 2003-06-16 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of the Respiratory Chain Inhibitor Antimycin to the Mitochondrial bc(1) Complex: A New Crystal Structure Reveals an Altered Intramolecular Hydrogen-bonding Pattern.
J.Mol.Biol., 351, 2005
|
|
6IBC
 
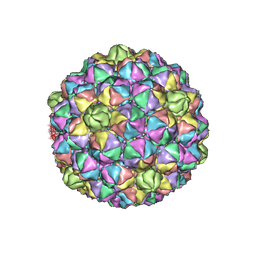 | | Thermophage P23-45 procapsid | | Descriptor: | Major head protein | | Authors: | Bayfield, O.W, Klimuk, E, Winkler, D.C, Hesketh, E.L, Chechik, M, Cheng, N, Dykeman, E.C, Minakhin, L, Ranson, N.A, Severinov, K, Steven, A.C, Antson, A.A. | | Deposit date: | 2018-11-29 | | Release date: | 2019-02-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5XPL
 
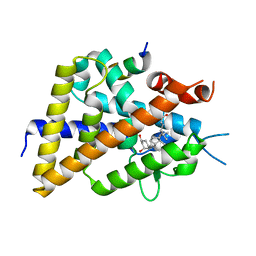 | | Crystal structure of VDR-LBD complexed with 22S-butyl-25-hydroxyphenyl-2-methylidene-19,26,27-trinor-25-oxo-1-hydroxyvitamin D3 | | Descriptor: | (4~{S})-4-[(1~{R})-1-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[2-[(3~{R},5~{R})-4-methylidene-3,5-bis(oxidanyl )cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]ethyl]-1-(4-hydroxyphenyl)octan-1-one, Nuclear receptor coactivator 2, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
5XPM
 
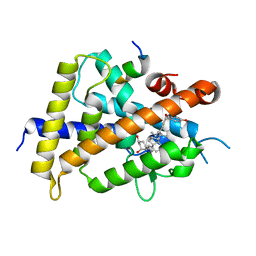 | | Crystal structure of VDR-LBD complexed with 22S-Butyl-25RS-(hydroxyphenyl)-25-methoxy-2-methylidene-19,26,27-trinor-1-hydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},3~{S})-3-[(3~{S})-3-(4-hydroxyphenyl)-3-methoxy-propyl]heptan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
7XT3
 
 | | Crystal Structure of Hepatitis virus A 2C protein 128-335 aa | | Descriptor: | Genome polyprotein, PHOSPHATE ION | | Authors: | Chen, P, Wojdyla, J.A, Li, Z, Wang, M, Cui, S. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Biochemical and structural characterization of hepatitis A virus 2C reveals an unusual ribonuclease activity on single-stranded RNA.
Nucleic Acids Res., 50, 2022
|
|
4A92
 
 | | Full-length HCV NS3-4A protease-helicase in complex with a macrocyclic protease inhibitor. | | Descriptor: | (1'R,2R,2'S,6S,24AS)-17-FLUORO-6-(1-METHYL-2-OXOPIPERIDINE-3-CARBOXAMIDO)-19,19-DIOXIDO-5,21,24-TRIOXO-2'-VINYL-1,2,3,5,6,7,8,9,10,11,12,13,14,20,21,23,24,24A-OCTADECAHYDROSPIRO[BENZO[S]PYRROLO[2,1-G][1,2,5,8,18]THIATETRAAZACYCLOICOSINE-22,1'-CYCLOPRO-2-CARBOXYLATEPAN]-2-YL 4-FLUOROISOINDOLINE, SERINE PROTEASE NS3, ZINC ION | | Authors: | Schiering, N, D'Arcy, A, Simic, O, Eder, J, Raman, P, Svergun, D.I, Bodendorf, U. | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A Macrocyclic Hcv Ns3/4A Protease Inhibitor Interacts with Protease and Helicase Residues in the Complex with its Full- Length Target.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3HNX
 
 | |
5XPP
 
 | | Crystal structure of VDR-LBD complexed with 25RS-(Hydroxyphenyl)-2-methylidene-19,26,27-trinor-1,25-dihydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{R})-6-(4-hydroxyphenyl)-6-oxidanyl-hexan-2-yl]-7~{ a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
3HNU
 
 | |
5WJE
 
 | | Crystal structure of Naa80 bound to a bisubstrate analogue | | Descriptor: | Actin N-terminus peptide, CARBOXYMETHYL COENZYME *A, CG8481, ... | | Authors: | Goris, M, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2017-07-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | Structural determinants and cellular environment define processed actin as the sole substrate of the N-terminal acetyltransferase NAA80.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1N6Z
 
 | |
1NB5
 
 | | Crystal structure of stefin A in complex with cathepsin H | | Descriptor: | Cathepsin H, Cathepsin H MINI CHAIN, STEFIN A, ... | | Authors: | Jenko, S, Dolenc, I, Guncar, G, Dobersek, A, Podobnik, M, Turk, D. | | Deposit date: | 2002-12-02 | | Release date: | 2003-02-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo- and exopeptidases
J.Mol.Biol., 326, 2003
|
|
6JW7
 
 | | The crystal structure of KanD2 in complex with NADH and 3"-deamino-3"-hydroxykanamycin A | | Descriptor: | (2R,3S,4S,5R,6R)-2-(aminomethyl)-6-[(1R,2S,3S,4R,6S)-4,6-bis(azanyl)-3-[(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-2-oxidanyl-cyclohexyl]oxy-oxane-3,4,5-triol, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Dehydrogenase | | Authors: | Kudo, F, Kitayama, Y, Miyanaga, A, Hirayama, A, Eguchi, T. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Biochemical and structural analysis of a dehydrogenase, KanD2, and an aminotransferase, KanS2, that are responsible for the construction of the kanosamine moiety in kanamycin biosynthesis.
Biochemistry, 59, 2020
|
|
5WEY
 
 | | Joint X-ray/neutron structure of Concanavalin A with alpha1-2 D-mannobiose | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Kovalevsky, A, Gerlits, O.O, Woods, R.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | Mannobiose Binding Induces Changes in Hydrogen Bonding and Protonation States of Acidic Residues in Concanavalin A As Revealed by Neutron Crystallography.
Biochemistry, 56, 2017
|
|
5WKA
 
 | | Crystal structure of a GH1 beta-glucosidase retrieved from microbial metagenome of Poraque Amazon lake | | Descriptor: | Beta-glucosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Morais, M.A.B, Toyama, D, Ramos, F.C, Zanphorlin, L.M, Tonoli, C.C.C, Miranda, F.P, Ruller, R, Henrique-Silva, F, Murakami, M.T. | | Deposit date: | 2017-07-24 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A novel beta-glucosidase isolated from the microbial metagenome of Lake Poraque (Amazon, Brazil).
Biochim. Biophys. Acta, 1866, 2018
|
|
4YY6
 
 | | Crystal structure of BRD9 Bromodomain bound to a butyryllysine peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
4YYD
 
 | | Crystal structure of BRD9 Bromodomain bound to a crotonyllysine peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
4ATZ
 
 | |
2VH4
 
 | | Structure of a loop C-sheet serpin polymer | | Descriptor: | TENGPIN | | Authors: | Zhang, Q, Law, R.H.P, Bottomley, S.P, Whisstock, J.C, Buckle, A.M. | | Deposit date: | 2007-11-19 | | Release date: | 2008-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Structural Basis for Loop C-Sheet Polymerization in Serpins.
J.Mol.Biol., 376, 2008
|
|
1K8A
 
 | | Co-crystal structure of Carbomycin A bound to the 50S ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T. | | Deposit date: | 2001-10-23 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
