4I10
 
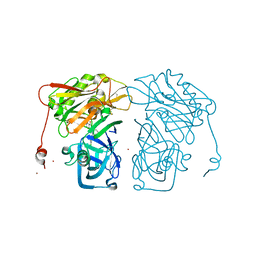 | | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates | | Descriptor: | 2-{(1S)-1-[(6-chloro-3,3-dimethyl-3,4-dihydroisoquinolin-1-yl)amino]-2-phenylethyl}pyrido[4,3-d]pyrimidin-4(1H)-one, Beta-secretase 1, ZINC ION | | Authors: | Lougheed, J.C, Brecht, E, Yao, N.H. | | Deposit date: | 2012-11-19 | | Release date: | 2013-03-06 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3FH9
 
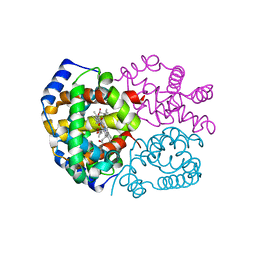 | | Crystal structure determination of indian flying fox (Pteropus giganteus) at 1.62 A resolution | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, OXYGEN MOLECULE, ... | | Authors: | Sathya Moorthy, Pon, Neelagandan, K, Balasubramanian, M, Thenmozhi, M, Ponnuswamy, M.N. | | Deposit date: | 2008-12-09 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure determination of indian flying fox (Pteropus giganteus) at 1.62 A resolution
To be Published
|
|
4EIH
 
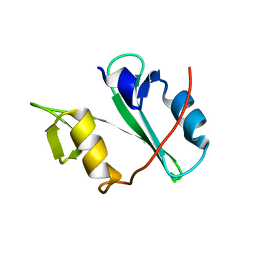 | | Crystal structure of Arg SH2 domain | | Descriptor: | Abelson tyrosine-protein kinase 2, CHLORIDE ION | | Authors: | Liu, W, MacGrath, S.M, Koleske, A.J, Boggon, T.J. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Two Amino Acid Residues Confer Different Binding Affinities of Abelson Family Kinase Src Homology 2 Domains for Phosphorylated Cortactin.
J.Biol.Chem., 289, 2014
|
|
4EJL
 
 | |
4ENH
 
 | |
3FJ5
 
 | | Crystal structure of the c-src-SH3 domain | | Descriptor: | ACETATE ION, GLYCEROL, Proto-oncogene tyrosine-protein kinase Src, ... | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-12-14 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Intertwined dimeric structure for the SH3 domain of the c-Src tyrosine kinase induced by polyethylene glycol binding
Febs Lett., 583, 2009
|
|
3FF9
 
 | | Structure of NK cell receptor KLRG1 | | Descriptor: | Killer cell lectin-like receptor subfamily G member 1 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
4HY9
 
 | |
3ZS2
 
 | | TyrB25,NMePheB26,LysB28,ProB29-insulin analogue crystal structure | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Antolikova, E, Zakova, L, Turkenburg, J.P, Watson, C.J, Hanclova, I, Sanda, M, Cooper, A, Kraus, T, Brzozowski, A.M, Jiracek, J.A. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Non-Equivalent Role of Inter- and Intramolecular Hydrogen Bonds in the Insulin Dimer Interface.
J.Biol.Chem., 286, 2011
|
|
4ET9
 
 | | Hen egg-white lysozyme solved from 5 fs free-electron laser pulse data | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4L05
 
 | | Cu/Zn superoxide dismutase from Brucella abortus | | Descriptor: | COPPER (I) ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Shin, D.S, Didonato, M, Pratt, A.J, Bruns, C.K, Cabelli, D.E, Kroll, J.S, Belzer, C.A, Tabatabai, L.B, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2013-05-30 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.098 Å) | | Cite: | Structural, Functional, and Immunogenic Insights on Cu,Zn Superoxide Dismutase Pathogenic Virulence Factors from Neisseria meningitidis and Brucella abortus.
J.Bacteriol., 197, 2015
|
|
4ETD
 
 | | Lysozyme, room-temperature, rotating anode, 0.0026 MGy | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4L0F
 
 | |
3ZOY
 
 | | Crystal Structure of N64Del Mutant of Nitrosomonas europaea Cytochrome c552 (hexagonal space group) | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
3ZQ3
 
 | | Crystal Structure of Rat Odorant Binding Protein 3 (OBP3) | | Descriptor: | OBP3 PROTEIN | | Authors: | Portman, K.L, Long, J, Carr, S, Brand, L, Winzor, D.J, Searle, M, Scott, D.J. | | Deposit date: | 2013-03-05 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enthalpy/Entropy Compensation Effects from Cavity Desolvation Underpin Broad Ligand Binding Selectivity for Rat Odorant Binding Protein 3
Biochemistry, 53, 2014
|
|
4L36
 
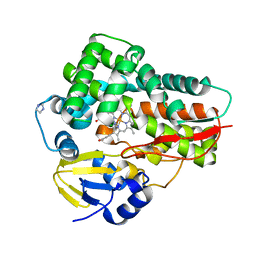 | | Crystal structure of the cytochrome P450 enzyme TxtE | | Descriptor: | HEME B/C, IMIDAZOLE, Putative P450-like protein | | Authors: | Yu, F, Li, M.J, Xu, C.Y, Wang, Z.J, Zhou, H, Yang, M, Chen, Y.X, Tang, L, He, J.H. | | Deposit date: | 2013-06-05 | | Release date: | 2013-12-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural Insights into the Mechanism for Recognizing Substrate of the Cytochrome P450 Enzyme TxtE.
Plos One, 8, 2013
|
|
3F02
 
 | |
4L4A
 
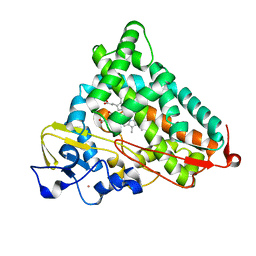 | | Structure of L358A/K178G mutant of P450cam bound to camphor | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Batabyal, D, Li, H, Poulos, T.L. | | Deposit date: | 2013-06-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Synergistic Effects of Mutations in Cytochrome P450cam Designed To Mimic CYP101D1.
Biochemistry, 52, 2013
|
|
4L54
 
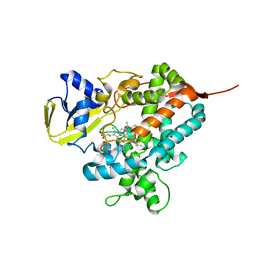 | | Structure of cytochrome P450 OleT, ligand-free | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Terminal olefin-forming fatty acid decarboxylase | | Authors: | Leys, D. | | Deposit date: | 2013-06-10 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Biochemical Properties of the Alkene Producing Cytochrome P450 OleTJE (CYP152L1) from the Jeotgalicoccus sp. 8456 Bacterium.
J.Biol.Chem., 289, 2014
|
|
4L85
 
 | |
3FO7
 
 | | Simultaneous inhibition of anti-coagulation and inflammation: Crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand binding site | | Descriptor: | INDOMETHACIN, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Prem Kumar, R, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2008-12-29 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Simultaneous inhibition of anti-coagulation and inflammation: Crystal structure of phospholipase A2 complexed with indomethacin at 1.4 A resolution reveals the presence of the new common ligand binding site
To be Published
|
|
4E7T
 
 | | The structure of T6 bovine insulin | | Descriptor: | Insulin A chain, Insulin B chain, ZINC ION | | Authors: | Harris, P, Frankaer, C.G, Knudsen, M.V. | | Deposit date: | 2012-03-19 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structures of T(6), T(3)R(3) and R(6) bovine insulin: combining X-ray diffraction and absorption spectroscopy.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3F3R
 
 | | Crystal structure of yeast Thioredoxin1-glutathione mixed disulfide complex | | Descriptor: | GLUTATHIONE, SULFATE ION, Thioredoxin-1 | | Authors: | Zhang, Y.R, Bao, R, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2008-10-31 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic analysis of Saccharomyces cerevisiae thioredoxin Trx1: implications for the catalytic mechanism of GSSG reduced by the thioredoxin system
Biochim.Biophys.Acta, 1794, 2009
|
|
3F6C
 
 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI | | Descriptor: | GLYCEROL, Positive transcription regulator evgA | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Wu, B, Bain, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI
To be Published
|
|
4EFG
 
 | |
