3F1K
 
 | |
3FE3
 
 | |
3F3W
 
 | | Drug resistant cSrc kinase domain in complex with inhibitor RL45 (Type II) | | Descriptor: | 1-{4-[(6-aminoquinazolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(3-methylphenyl)-1H-pyrazol-5-yl]urea, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Grutter, C, Kluter, S, Getlik, M, Rauh, D. | | Deposit date: | 2008-10-31 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hybrid compound design to overcome the gatekeeper T338M mutation in cSrc
J.Med.Chem., 52, 2009
|
|
3FG1
 
 | | Crystal structure of Delta413-417:GS LOX | | Descriptor: | ACETIC ACID, Allene oxide synthase-lipoxygenase protein, CALCIUM ION, ... | | Authors: | Neau, D.B, Newcomer, M.E. | | Deposit date: | 2008-12-04 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A structure of an 8R-lipoxygenase suggests a general model for lipoxygenase product specificity.
Biochemistry, 48, 2009
|
|
3F6P
 
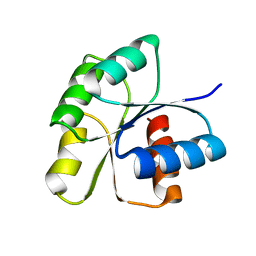 | | Crystal Structure of unphosphorelated receiver domain of YycF | | Descriptor: | Transcriptional regulatory protein yycF | | Authors: | Zhao, H, Tang, L. | | Deposit date: | 2008-11-06 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Preliminary crystallographic studies of the regulatory domain of response regulator YycF from an essential two-component signal transduction system.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3FJS
 
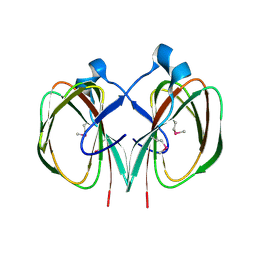 | |
3F7J
 
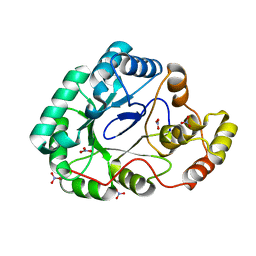 | | B.subtilis YvgN | | Descriptor: | NITRATE ION, POTASSIUM ION, YvgN protein | | Authors: | Zhou, Y.F, Lei, J, Liang, Y.H, Su, X.-D. | | Deposit date: | 2008-11-09 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analyses of YvgN and YtbE from Bacillus subtilis
Protein Sci., 18, 2009
|
|
3F7Z
 
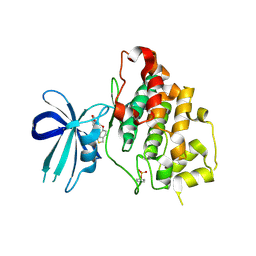 | | X-ray Co-Crystal Structure of Glycogen Synthase Kinase 3beta in Complex with an Inhibitor | | Descriptor: | 2-(1,3-benzodioxol-5-yl)-5-[(3-fluoro-4-methoxybenzyl)sulfanyl]-1,3,4-oxadiazole, Glycogen synthase kinase-3 beta | | Authors: | Mol, C.D, Dougan, D.R. | | Deposit date: | 2008-11-10 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design, synthesis and structure-activity relationships of 1,3,4-oxadiazole derivatives as novel inhibitors of glycogen synthase kinase-3beta.
Bioorg.Med.Chem., 17, 2009
|
|
3F9T
 
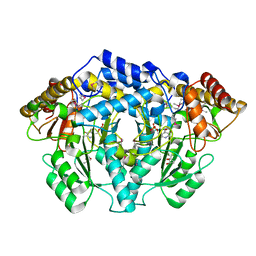 | |
3FOO
 
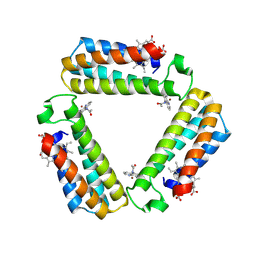 | |
3FB3
 
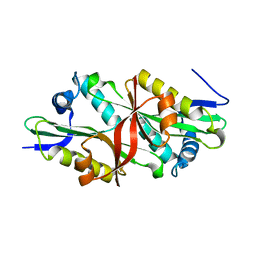 | | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886 | | Descriptor: | N-acetyltransferase | | Authors: | Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, MacKenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-18 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886
TO BE PUBLISHED
|
|
3FC2
 
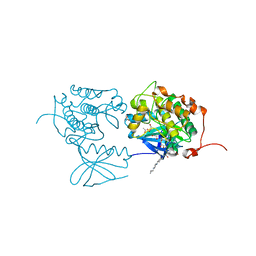 | | PLK1 in complex with BI6727 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bader, G. | | Deposit date: | 2008-11-21 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | BI 6727, A Polo-like Kinase Inhibitor with Improved Pharmacokinetic Profile and Broad Antitumor Activity.
Clin.Cancer Res., 15, 2009
|
|
3FD4
 
 | |
3FD7
 
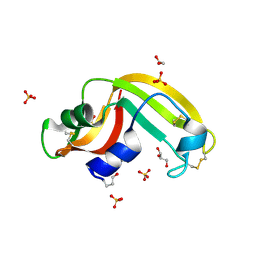 | | Crystal structure of Onconase C87A/C104A-ONC | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein P-30, ... | | Authors: | Neumann, P, Schulenburg, C, Arnold, U, Ulbrich-Hofmann, R, Stubbs, M.T. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Impact of the C-terminal disulfide bond on the folding and stability of onconase.
Chembiochem, 11, 2010
|
|
3EXB
 
 | |
3EXL
 
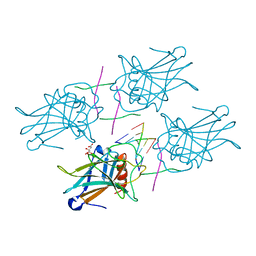 | | Crystal Structure of a p53 Core Tetramer Bound to DNA | | Descriptor: | 5'-D(*DGP*DAP*DGP*DCP*DAP*DTP*DGP*DCP*DTP*DCP*DA)-3', 5'-D(*DTP*DTP*DGP*DAP*DGP*DCP*DAP*DTP*DGP*DCP*DTP*DC)-3', CITRATE ANION, ... | | Authors: | Malecka, K.A. | | Deposit date: | 2008-10-16 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a p53 core tetramer bound to DNA.
Oncogene, 28, 2009
|
|
3EXS
 
 | | Crystal structure of KGPDC from Streptococcus mutans in complex with D-R5P | | Descriptor: | RIBULOSE-5-PHOSPHATE, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Wang, K.T, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3FFR
 
 | |
3F3M
 
 | |
3FG6
 
 | | Structure of the C-terminus of Adseverin | | Descriptor: | Adseverin, CALCIUM ION | | Authors: | Robinson, R.C. | | Deposit date: | 2008-12-05 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the C-terminus of adseverin reveals the actin-binding interface.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3F4R
 
 | | Crystal structure of Wolbachia pipientis alpha-DsbA1 | | Descriptor: | PENTAETHYLENE GLYCOL, Putative uncharacterized protein, TRIETHYLENE GLYCOL | | Authors: | Kurz, M, Heras, B, Martin, J.L. | | Deposit date: | 2008-11-02 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of the Oxidoreductase alpha-DsbA1 from Wolbachia pipientis
ANTIOXID.REDOX SIGNAL., 11, 2009
|
|
3FLR
 
 | |
3F7X
 
 | |
3EVT
 
 | | Crystal structure of phosphoglycerate dehydrogenase from Lactobacillus plantarum | | Descriptor: | Phosphoglycerate dehydrogenase | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Do, J, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of phosphoglycerate dehydrogenase from Lactobacillus plantarum
To be Published
|
|
3EWK
 
 | |
