7DLX
 
 | | crystal structure of H2AM4>Z-H2B | | Descriptor: | Histone H2B,Histone H2A | | Authors: | Dai, L.C, Zhou, Z. | | Deposit date: | 2020-11-30 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Recognition of the inherently unstable H2A nucleosome by Swc2 is a major determinant for unidirectional H2A.Z exchange.
Cell Rep, 35, 2021
|
|
5A7I
 
 | | Crystal structure of INPP5B in complex with biphenyl 3,3',4,4',5,5'- hexakisphosphate | | Descriptor: | Biphenyl 3,3',4,4',5,5'-hexakisphosphate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Mills, S.J, Silvander, C, Cozier, G, Potter, B.V.L, Norldund, P. | | Deposit date: | 2015-07-06 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal Structures of Type-II Inositol Polyphosphate 5-Phosphatase Inpp5B with Synthetic Inositol Polyphosphate Surrogates Reveal New Mechanistic Insights for the Inositol 5-Phosphatase Family.
Biochemistry, 55, 2016
|
|
4ZSQ
 
 | | BACE crystal structure with tricyclic aminothiazine inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-[(4S,4aS,6S,8aR)-10-aminohexahydro-3H-4,8a-(epithiomethenoazeno)isochromen-6(1H)-yl]-3-chlorobenzamide | | Authors: | Timm, D.E. | | Deposit date: | 2015-05-13 | | Release date: | 2015-06-10 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Preparation and biological evaluation of conformationally constrained BACE1 inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
6E9U
 
 | | The crystal structure of bovine ultralong antibody BOV-7 | | Descriptor: | Bovine ultralong antibody BOV-7 heavy chain, Bovine ultralong antibody BOV-7 light chain | | Authors: | Dong, J, Crowe, J.E. | | Deposit date: | 2018-08-01 | | Release date: | 2019-05-01 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Structural Diversity of Ultralong CDRH3s in Seven Bovine Antibody Heavy Chains.
Front Immunol, 10, 2019
|
|
8EZB
 
 | | NHEJ Long-range complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (30-MER), DNA (31-MER), ... | | Authors: | Chen, S, He, Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ.
Sci Adv, 9, 2023
|
|
8EZA
 
 | | NHEJ Long-range complex with PAXX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (30-MER), DNA (31-MER), ... | | Authors: | Chen, S, He, Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ.
Sci Adv, 9, 2023
|
|
4ZYB
 
 | | High resolution structure of M23 peptidase LytM with substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Grabowska, M, Jagielska, E, Czapinska, H, Bochtler, M, Sabala, I. | | Deposit date: | 2015-05-21 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution structure of an M23 peptidase with a substrate analogue.
Sci Rep, 5, 2015
|
|
4ZZ8
 
 | | X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Glucanase/chitosanase, ... | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
4CTD
 
 | | X-ray structure of an engineered OmpG loop6-deletion | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CHLORIDE ION, OUTER MEMBRANE PROTEIN G | | Authors: | Grosse, W, Essen, L.-O. | | Deposit date: | 2014-03-13 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Based Engineering of a Minimal Porin Reveals Loop- Independent Channel Closure.
Biochemistry, 53, 2014
|
|
7DBN
 
 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V/F160M:DNA:dCTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
7DFC
 
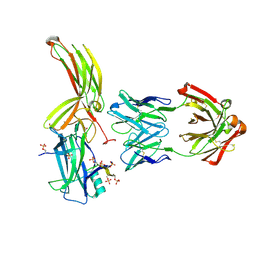 | | Crystal of Arrestin2-V2Rpp-3-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
4D7Y
 
 | | Crystal structure of mouse C1QL1 globular domain | | Descriptor: | C1Q-RELATED FACTOR, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kakegawa, W, Mitakidis, N, Miura, E, Abe, M, Matsuda, K, Takeo, Y, Kohda, K, Motohashi, J, Takahashi, A, Nagao, S, Muramatsu, S, Watanabe, M, Sakimura, K, Aricescu, A.R, Yuzaki, M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Anterograde C1Ql1 Signaling is Required in Order to Determine and Maintain a Single-Winner Climbing Fiber in the Mouse Cerebellum
Neuron, 85, 2015
|
|
5AA3
 
 | |
4D60
 
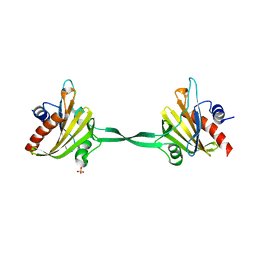 | | Structure of a dimeric Plasmodium falciparum profilin mutant | | Descriptor: | PROFILIN, SULFATE ION | | Authors: | Bhargav, S.P, Vahokoski, J, Kallio, J.P, Torda, A, Kursula, P, Kursula, I. | | Deposit date: | 2014-11-07 | | Release date: | 2015-06-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Two Independently Folding Units of Plasmodium Profilin Suggest Evolution Via Gene Fusion.
Cell.Mol.Life Sci., 72, 2015
|
|
7DBM
 
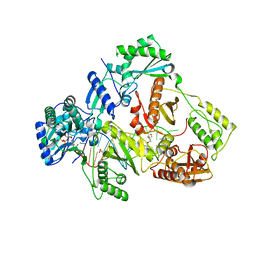 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
5A80
 
 | | Crystal structure of human JMJD2A in complex with compound 40 | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-[2-(3-methoxyphenyl)ethanoylamino]-2-oxidanyl-phenyl]pyridine-4-carboxylic acid, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Kopec, J, Tallant, C, Froese, S, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-11 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
8F3C
 
 | | Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (38-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2022-11-09 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5AC8
 
 | | S. enterica HisA with mutations D10G, dup13-15, G102A | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, SULFATE ION | | Authors: | Newton, M, Guo, X, Soderholm, A, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2015-08-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5A2M
 
 | | Thrombin Inhibitor | | Descriptor: | (2S)-1-[(2R)-5-carbamimidamido-2-[(phenylmethyl)sulfonylamino]pentanoyl]-N-[[5-chloranyl-2-(hydroxymethyl)phenyl]methyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2015-05-20 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Thrombin Inhibition
To be Published
|
|
5A2T
 
 | | The Molecular Basis for Flexibility in the Flexible Filamentous Plant Viruses | | Descriptor: | BAMBOO MOSAIC VIRUS, COAT PROTEIN | | Authors: | DiMaio, F, Chen, C.C, Yu, X, Frenz, B, Hsu, Y.H, Lin, N.S, Egelman, E.H. | | Deposit date: | 2015-05-23 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | The Molecular Basis for Flexibility in the Flexible Filamentous Plant Viruses.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4CX8
 
 | | Monomeric pseudorabies virus protease pUL26N at 2.5 A resolution | | Descriptor: | PSEUDORABIES VIRUS PROTEASE | | Authors: | Zuehlsdorf, M, Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-04-04 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin Pul26N, a Herpesvirus Serine Protease.
Plos Pathog., 11, 2015
|
|
8EEV
 
 | | Venezuelan equine encephalitis virus-like particle in complex with Fab SKT-20 | | Descriptor: | Coat protein, Fab SKT20 heavy chain, Fab SKT20 light chain | | Authors: | Tsybovsky, Y, Pletnev, S, Verardi, R, Roederer, M, Kwong, P.D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
6E7D
 
 | | Structure of the inhibitory NKR-P1B receptor bound to the host-encoded ligand, Clr-b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D, Killer cell lectin-like receptor subfamily B member 1B allele B, ... | | Authors: | Balaji, G.R, Rossjohn, J, Berry, R. | | Deposit date: | 2018-07-26 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition of host Clr-b by the inhibitory NKR-P1B receptor provides a basis for missing-self recognition.
Nat Commun, 9, 2018
|
|
5ACW
 
 | | VIM-2-1, Discovery of novel inhibitor scaffolds against the metallo- beta-lactamase VIM-2 by SPR based fragment screening | | Descriptor: | 4-methyl-5-(trifluoromethyl)-1,2,4-triazole-3-thiol, BETA-LACTAMASE, CHLORIDE ION, ... | | Authors: | Christopeit, T, Carlsen, T.J.O, Helland, R, Leiros, H.K.S. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Novel Inhibitor Scaffolds Against the Metallo-Beta-Lactamase Vim-2 by Spr Based Fragment Screening
J.Med.Chem., 58, 2015
|
|
3W07
 
 | |
