7DYD
 
 | |
7DX4
 
 | | The structure of FC08 Fab-hA.CE2-RBD complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Heavy chain of FC08 Fab, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2021-01-18 | | Release date: | 2021-04-21 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A proof of concept for neutralizing antibody-guided vaccine design against SARS-CoV-2.
Natl Sci Rev, 8, 2021
|
|
7DK3
 
 | | SARS-CoV-2 S trimer, S-open | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-16 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7E8M
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with mutated RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | Authors: | Wang, X.Q, Zhang, L.Q, Ge, J.W, Wang, R.K, Lan, J. | | Deposit date: | 2021-03-02 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species.
Immunity, 54, 2021
|
|
7DZY
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 2490 | | Descriptor: | Fab Heavy chain of enhancing antibody 2490, Fab light chain of enhancing antibody 2490, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7DZX
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 8D2 | | Descriptor: | Fab Heavy chain of enhancing antibody, Fab light chain of enhancing antibody, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7DZW
 
 | | Apo spike protein from SARS-CoV2 | | Descriptor: | Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7EK6
 
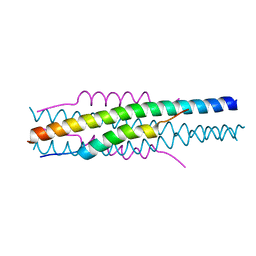 | | Structure of viral peptides IPB19/N52 | | Descriptor: | Spike protein S2 | | Authors: | Yu, D, Qin, B, Cui, S, He, Y. | | Deposit date: | 2021-04-04 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.243 Å) | | Cite: | Structure-based design and characterization of novel fusion-inhibitory lipopeptides against SARS-CoV-2 and emerging variants.
Emerg Microbes Infect, 10, 2021
|
|
7EAZ
 
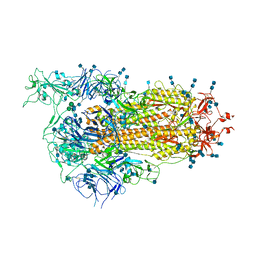 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, one RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7E5Y
 
 | | Molecular basis for neutralizing antibody 2B11 targeting SARS-CoV-2 RBD | | Descriptor: | 2B11 Fab Heavy chain, 2B11 Fab Light chain, Spike protein S1 | | Authors: | Wu, H, Yu, F, Wang, Q.S, Zhou, H, Wang, W.W, Zhao, T, Pan, Y.B, Yang, X.M. | | Deposit date: | 2021-02-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Screening of potent neutralizing antibodies against SARS-CoV-2 using convalescent patients-derived phage-display libraries.
Cell Discov, 7, 2021
|
|
7EB5
 
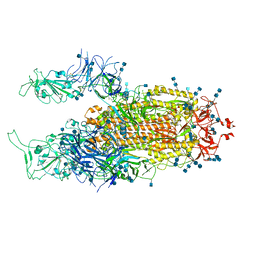 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, two RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7EB3
 
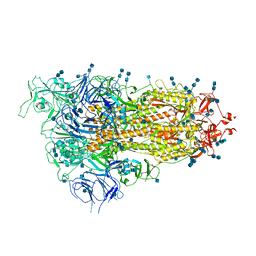 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, one RBD-up conformation 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7EB0
 
 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, one RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7EB4
 
 | | Cryo-EM structure of SARS-CoV-2 Spike D614G variant, two RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-03-08 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | D614G mutation in the SARS-CoV-2 spike protein enhances viral fitness by desensitizing it to temperature-dependent denaturation.
J.Biol.Chem., 297, 2021
|
|
7D4G
 
 | |
7DPV
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-dihydromyricetin | | Descriptor: | (2S,3S)-3,5-dihydroxy-7-methoxy-2-(3,4,5-trihydroxyphenyl)chroman-4-one, 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DPP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DPU
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-myricetin | | Descriptor: | 3C-like proteinase, 7-methoxy-3,5-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one, GLYCEROL | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DDP
 
 | | Cryo-EM structure of human ACE2 and GX/P2V/2017 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Niu, S, Wang, J, Wang, H.W, Qi, J.X, Wang, Q.H, Gao, G.F. | | Deposit date: | 2020-10-29 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin.
Embo J., 40, 2021
|
|
7DDO
 
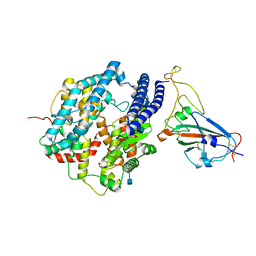 | | Cryo-EM structure of human ACE2 and GD/1/2019 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Niu, S, Wang, J, Wang, H.W, Qi, J.X, Wang, Q.H, Gao, G.F. | | Deposit date: | 2020-10-29 | | Release date: | 2021-05-19 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin.
Embo J., 40, 2021
|
|
7D2Z
 
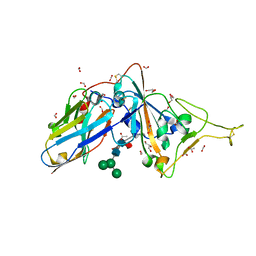 | | Structure of sybody SR31 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A high-affinity RBD-targeting nanobody improves fusion partner's potency against SARS-CoV-2.
Plos Pathog., 17, 2021
|
|
7D30
 
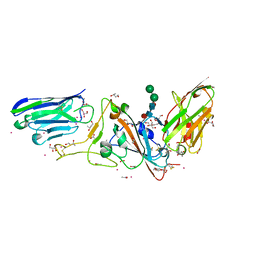 | | Structure of sybody MR17-SR31 fusion in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 1,3-BUTANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A high-affinity RBD-targeting nanobody improves fusion partner's potency against SARS-CoV-2.
Plos Pathog., 17, 2021
|
|
7CZP
 
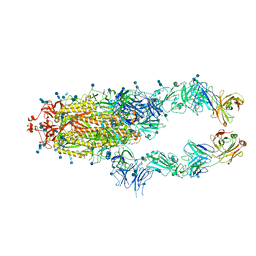 | | S protein of SARS-CoV-2 in complex bound with P2B-1A1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c181_light_IGLV2-14_IGLJ3,IGL@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZQ
 
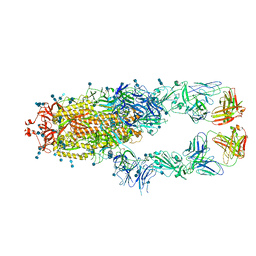 | | S protein of SARS-CoV-2 in complex bound with P2B-1A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c642_heavy_IGHV3-53_IGHD1-26_IGHJ6,Chain H of P2B-1A10,Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZR
 
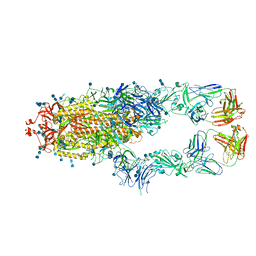 | | S protein of SARS-CoV-2 in complex bound with P5A-1B8_2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c542_heavy_IGHV3-53_IGHD3-10_IGHJ6,IGH@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
