7JV8
 
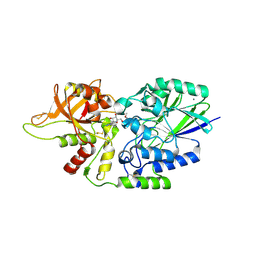 | | Human CD73 (ecto 5'-nucleotidase) in complex with compound 35 | | Descriptor: | 5'-nucleotidase, 6-chloro-N-cyclopentyl-1-{5-O-[(2R)-1-hydroxy-3-methoxy-2-phosphonopropan-2-yl]-beta-D-ribofuranosyl}-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALCIUM ION, ... | | Authors: | Gibbons, P, Du, X. | | Deposit date: | 2020-08-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Orally Bioavailable Small-Molecule CD73 Inhibitor (OP-5244) Reverses Immunosuppression through Blockade of Adenosine Production.
J.Med.Chem., 63, 2020
|
|
5AX0
 
 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.52 angstrom | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | Authors: | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5AWZ
 
 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.57 angstrom | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | Authors: | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5AX1
 
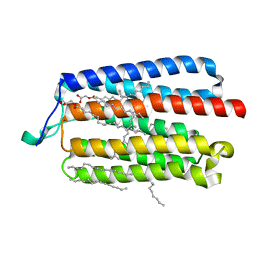 | | Crystal Structure of the Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, at 1.80 angstrom | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DECANE, DODECANE, ... | | Authors: | Furuse, M, Hosaka, T, Kimura-Someya, T, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural basis for the slow photocycle and late proton release in Acetabularia rhodopsin I from the marine plant Acetabularia acetabulum
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5GAO
 
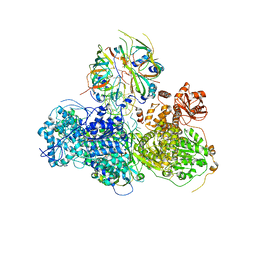 | | Head region of the yeast spliceosomal U4/U6.U5 tri-snRNP | | Descriptor: | Pre-mRNA-splicing factor 8, Pre-mRNA-splicing helicase BRR2, Saccharomyces cerevisiae strain UOA_M2 chromosome 5 sequence, ... | | Authors: | Nguyen, T.H.D, Galej, W.P, Bai, X.C, Oubridge, C, Scheres, S.H.W, Newman, A.J, Nagai, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
8GOE
 
 | | Structure of hSLC19A1+5-MTHF | | Descriptor: | N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-08-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
8GOF
 
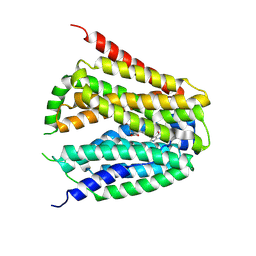 | | Structure of hSLC19A1+PMX | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-08-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
2BHD
 
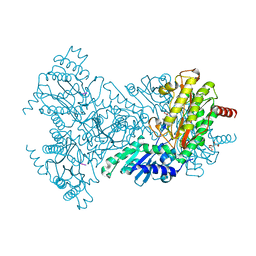 | | Mg substituted E. coli Aminopeptidase P in complex with product | | Descriptor: | CITRATE ANION, MAGNESIUM ION, VALINE-PROLINE-LEUCINE TRIPEPTIDE, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2005-01-10 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, a Metalloprotease with a Dinuclear Metal Center.
Biochemistry, 44, 2005
|
|
2BHA
 
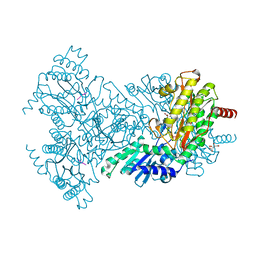 | | E. coli Aminopeptidase P in complex with substrate | | Descriptor: | CITRATE ANION, MAGNESIUM ION, VALINE-PROLINE-LEUCINE, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2005-01-08 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, a Metalloprotease with a Dinuclear Metal Center.
Biochemistry, 44, 2005
|
|
6MW0
 
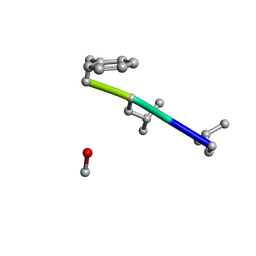 | | Mle-Phe-Mle-D-Phe. Linear tetrapeptide related to pseudoxylallemycin A. | | Descriptor: | METHANOL, Mle-Phe-Mle-D-Phe Linear tetrapeptide related to pseudoxylallemycin A | | Authors: | Cameron, A.J, Harris, P.W.R, Brimble, M.A, Squire, C.J. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Investigations of the key macrolactamisation step in the synthesis of cyclic tetrapeptide pseudoxylallemycin A.
Org.Biomol.Chem., 17, 2019
|
|
6MW1
 
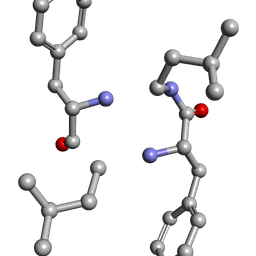 | |
6MW2
 
 | |
1TGJ
 
 | | HUMAN TRANSFORMING GROWTH FACTOR-BETA 3, CRYSTALLIZED FROM DIOXANE | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, TRANSFORMING GROWTH FACTOR-BETA 3 | | Authors: | Mittl, P.R.E, Priestle, J.P, Gruetter, M.G. | | Deposit date: | 1996-07-09 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of TGF-beta 3 and comparison to TGF-beta 2: implications for receptor binding.
Protein Sci., 5, 1996
|
|
6C3M
 
 | | Wild type structure of SiRHP | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Stroupe, M.E. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The role of extended Fe4S4cluster ligands in mediating sulfite reductase hemoprotein activity.
Biochim. Biophys. Acta, 1866, 2018
|
|
6C3Z
 
 | | T477A SiRHP | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Stroupe, M.E. | | Deposit date: | 2018-01-11 | | Release date: | 2018-06-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.681 Å) | | Cite: | The role of extended Fe4S4cluster ligands in mediating sulfite reductase hemoprotein activity.
Biochim. Biophys. Acta, 1866, 2018
|
|
6MVZ
 
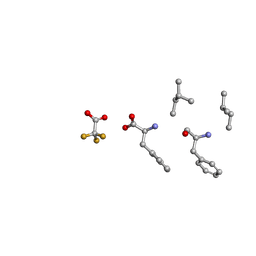 | | Mle-Phe-Mle-Phe. Linear precursor of pseudoxylallemycin A. | | Descriptor: | Linear precursor of pseudoxylallemycin A, trifluoroacetic acid | | Authors: | Cameron, A.J, Harris, P.W.R, Brimble, M.A, Squire, C.J. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | Investigations of the key macrolactamisation step in the synthesis of cyclic tetrapeptide pseudoxylallemycin A.
Org.Biomol.Chem., 17, 2019
|
|
6C3X
 
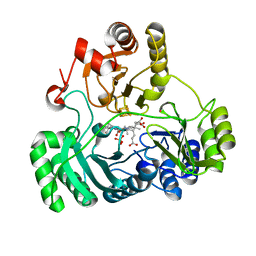 | | Wild type structure of SiRHP | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Stroupe, M.E. | | Deposit date: | 2018-01-11 | | Release date: | 2018-06-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.542 Å) | | Cite: | The role of extended Fe4S4cluster ligands in mediating sulfite reductase hemoprotein activity.
Biochim. Biophys. Acta, 1866, 2018
|
|
7XPZ
 
 | | Structure of Apo-hSLC19A1 | | Descriptor: | Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XQ0
 
 | | Structure of hSLC19A1+3'3'-CDA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XQ2
 
 | | Structure of hSLC19A1+2'3'-cGAMP | | Descriptor: | Reduced folate transporter, cGAMP | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XQ1
 
 | | Structure of hSLC19A1+2'3'-CDAS | | Descriptor: | (1~{R},3~{S},6~{R},8~{R},9~{R},10~{S},12~{S},15~{R},17~{R},18~{R})-8,17-bis(6-aminopurin-9-yl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecane-9,18-diol, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
6C3Y
 
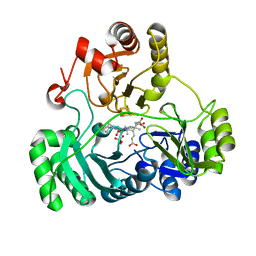 | | Wild type structure of SiRHP | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Stroupe, M.E. | | Deposit date: | 2018-01-11 | | Release date: | 2018-06-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.542 Å) | | Cite: | The role of extended Fe4S4cluster ligands in mediating sulfite reductase hemoprotein activity.
Biochim. Biophys. Acta, 1866, 2018
|
|
1DK6
 
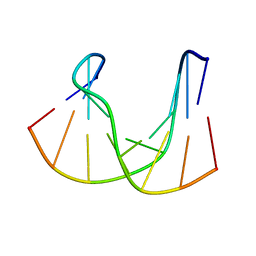 | | NMR structure analysis of the DNA nine base pair duplex D(CATGAGTAC) D(GTAC(NP3)CATG) | | Descriptor: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*(NP3)P*CP*AP*TP*GP*)-3' | | Authors: | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | Deposit date: | 1999-12-06 | | Release date: | 2000-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
1DK9
 
 | | SOLUTION STRUCTURE ANALYSIS OF THE DNA DUPLEX D(CATGAGTAC)D(GTACTCATG) | | Descriptor: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*TP*CP*AP*TP*GP*)-3' | | Authors: | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | Deposit date: | 1999-12-06 | | Release date: | 1999-12-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
1TGK
 
 | |
