3K31
 
 | |
8D2V
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 1B conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2U
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 1A conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2S
 
 | | Zebrafish MFSD2A isoform B in inward open ligand bound conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2W
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 2B conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
4C2V
 
 | | Aurora B kinase in complex with the specific inhibitor Barasertib | | Descriptor: | 2-[5-[[7-[3-[ethyl(2-hydroxyethyl)amino]propoxy]quinazolin-4-yl]amino]-1H-pyrazol-3-yl]-N-(3-fluorophenyl)ethanamide, AURORA KINASE B-A, INNER CENTROMERE PROTEIN A | | Authors: | Sessa, F, Villa, F. | | Deposit date: | 2013-08-20 | | Release date: | 2014-02-26 | | Last modified: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of Aurora B-Incenp in Complex with Barasertib Reveals a Potential Transinhibitory Mechanism
Acta Crystallogr.,Sect.F, 70, 2014
|
|
8D2T
 
 | | Zebrafish MFSD2A isoform B in inward open ligand-free conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2X
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 3C conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
6FM0
 
 | | Deoxyguanylosuccinate synthase (DgsS) and ATP structure at 1.7 Angstrom resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2018-01-29 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Science, 372, 2021
|
|
3I0O
 
 | | Crystal Structure of Spectinomycin Phosphotransferase, APH(9)-Ia, in complex with ADP and Spectinomcyin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Fong, D.H, Lemke, C.T, Hwang, J, Xiong, B, Berghuis, A.M. | | Deposit date: | 2009-06-25 | | Release date: | 2010-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila.
J.Biol.Chem., 285, 2010
|
|
4BWA
 
 | | PylRS Y306G, Y384F, I405R mutant in complex with adenylated norbornene | | Descriptor: | 1,2-ETHANEDIOL, Adenylated Norbornene, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schneider, S, Vrabel, M, Gattner, M.J, Fluegel, V, Lopez-Carillo, V, Carell, T. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Insights Into Incorporation of Norbornene Amino Acids for Click Modification of Proteins
Chem.Bio.Chem., 14, 2013
|
|
3GPO
 
 | | Crystal structure of macro domain of Chikungunya virus in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Malet, H, Jamal, S, Coutard, B, Canard, B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket
J.Virol., 83, 2009
|
|
4EEQ
 
 | | Crystal structure of E. faecalis DNA ligase with inhibitor | | Descriptor: | 4-amino-2-(cyclopentyloxy)pyrimidine-5-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Wang, T, Charifson, P, Wei, Y. | | Deposit date: | 2012-03-28 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis and Activity Evaluation of Potent NAD+ DNA Ligase Inhibitors as Potential Antibacterial Agents.
To be Published
|
|
3KDO
 
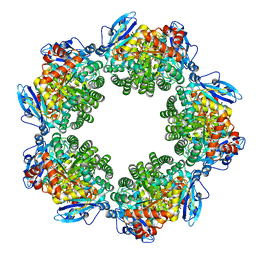 | | Crystal structure of Type III Rubisco SP6 mutant complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-10-23 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
3KDN
 
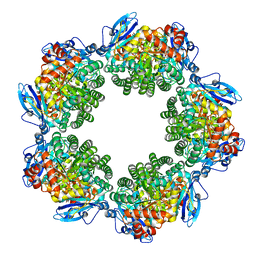 | | Crystal structure of Type III Rubisco SP4 mutant complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-10-23 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
3KLF
 
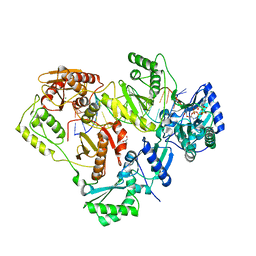 | | Crystal structure of wild-type HIV-1 Reverse Transcriptase crosslinked to a DSDNA with a bound excision product, AZTPPPPA | | Descriptor: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(2DA))-3'), DNA (5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), GLYCEROL, ... | | Authors: | Tu, X, Das, K, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3HGV
 
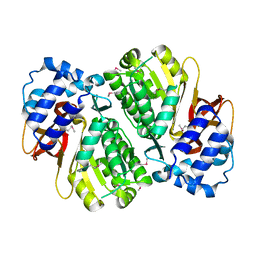 | |
4EMT
 
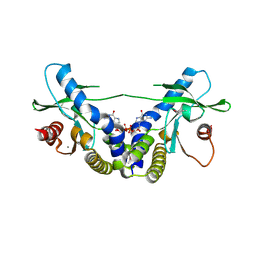 | | Crystal Structure of human STING bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Transmembrane protein 173 | | Authors: | Li, P. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-13 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3KLE
 
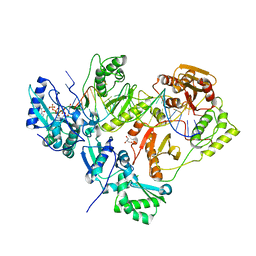 | | Crystal structure of AZT-resistant HIV-1 Reverse Transcriptase crosslinked to a DSDNA with a bound excision product, AZTPPPPA | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(2DA))-3'), GLYCEROL, ... | | Authors: | Tu, X, Das, K, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4DKX
 
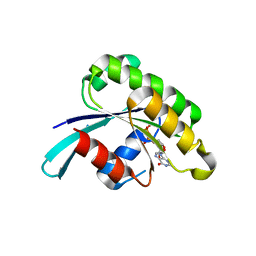 | | Crystal Structure of the Rab 6A'(Q72L) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-6A | | Authors: | Park, H.H, Shin, Y.-C. | | Deposit date: | 2012-02-04 | | Release date: | 2012-10-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Rab6A'(Q72L) mutant reveals unexpected GDP/Mg2+ binding with opened GTP-binding domain
Biochem.Biophys.Res.Commun., 424, 2012
|
|
3KLI
 
 | |
3L0P
 
 | | Crystal structures of Iron containing Adenylate kinase from Desulfovibrio gigas | | Descriptor: | Adenylate kinase, FE (III) ION, GLYCEROL | | Authors: | Mukhopadhyay, A, Trincao, J, Romao, M.J. | | Deposit date: | 2009-12-10 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the zinc-, cobalt-, and iron-containing adenylate kinase from Desulfovibrio gigas: a novel metal-containing adenylate kinase from Gram-negative bacteria
J.Biol.Inorg.Chem., 16, 2011
|
|
4EAT
 
 | | Crystal structure of a benzoate coenzyme A ligase | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, Benzoate-coenzyme A ligase, ... | | Authors: | Geiger, J, Strom, S. | | Deposit date: | 2012-03-22 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Kinetically and Crystallographically Guided Mutations of a Benzoate CoA Ligase (BadA) Elucidate Mechanism and Expand Substrate Permissivity.
Biochemistry, 54, 2015
|
|
4EHX
 
 | | Crystal structure of LpxK from Aquifex aeolicus at 1.9 angstrom resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Emptage, R.P, Daughtry, K.D, Pemble IV, C.W, Raetz, C.R.H. | | Deposit date: | 2012-04-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of LpxK, the 4'-kinase of lipid A biosynthesis and atypical P-loop kinase functioning at the membrane interface.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EFE
 
 | | crystal structure of DNA ligase | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, SULFATE ION, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of DNA ligase
To be Published
|
|
