3HX8
 
 | |
4U99
 
 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II) ligation state, Q154A/Q155A/K156A mutant | | Descriptor: | NO-binding heme-dependent sensor protein, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3AQT
 
 | | CRYSTAL STRUCTURE OF RolR (NCGL1110) complex WITH ligand RESORCINOL | | Descriptor: | Bacterial regulatory proteins, tetR family, RESORCINOL | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
3NRB
 
 | |
6HRQ
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with NCC-149 | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, Histone deacetylase, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-09-28 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HTI
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 6 | | Descriptor: | GLYCEROL, Histone deacetylase, POTASSIUM ION, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HTT
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 7 | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, Histone deacetylase, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
3AQS
 
 | | Crystal structure of RolR (NCGL1110) without ligand | | Descriptor: | Bacterial regulatory proteins, tetR family | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
6HT8
 
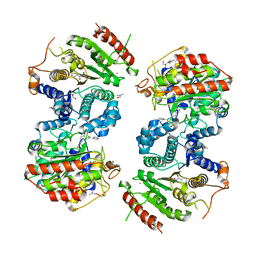 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 3 | | Descriptor: | 3-benzamido-4-methoxy-~{N}-oxidanyl-benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Marek, M, Shaik, T.B, Romier, C. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HU2
 
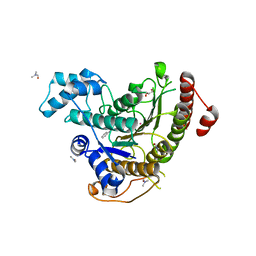 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 11 | | Descriptor: | 4-methyl-~{N}-oxidanyl-3-[(phenylmethyl)amino]benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
3NO4
 
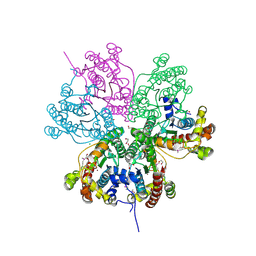 | |
3GWR
 
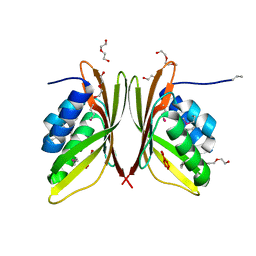 | |
3GPT
 
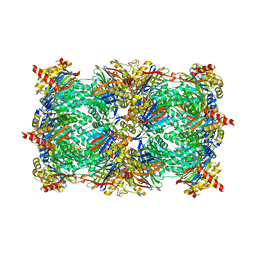 | | Crystal structure of the yeast 20S proteasome in complex with Salinosporamide derivatives: slow substrate ligand | | Descriptor: | (2R,3S,4R)-2-[(S)-(1S)-cyclohex-2-en-1-yl(hydroxy)methyl]-4-(2-fluoroethyl)-3-hydroxy-3-methyl-5-oxopyrrolidine-2-carbaldehyde, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Macherla, V.R, Manam, R.R, Arthur, K.A.M, Potts, C.B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Snapshots of the fluorosalinosporamide/20S complex offer mechanistic insights for fine tuning proteasome inhibition
J.Med.Chem., 52, 2009
|
|
3JTW
 
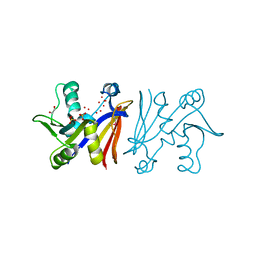 | |
6KAD
 
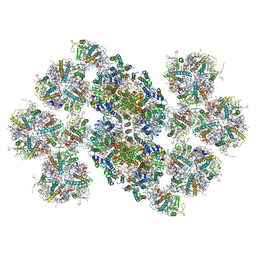 | | Cryo-EM structure of the C2S2M2L2-type PSII-LHCII supercomplex from Chlamydomonas reihardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Sheng, X, Watanabe, A, Li, A.J, Kim, E, Song, C, Murata, K, Song, D.F, Minagawa, J, Liu, Z.F. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insight into light harvesting for photosystem II in green algae.
Nat.Plants, 5, 2019
|
|
4HGD
 
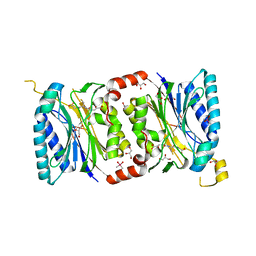 | | Structural insights into yeast Nit2: C169S mutant of yeast Nit2 in complex with an endogenous peptide-like ligand | | Descriptor: | CACODYLATE ION, GLYCEROL, N-(4-carboxy-4-oxobutanoyl)-L-cysteinylglycine, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-08 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6HSG
 
 | | Crystal structure of Schistosoma mansoni HDAC8 H292M mutant complexed with NCC-149 | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, Histone deacetylase, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
3H51
 
 | |
4GZV
 
 | |
6I8B
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor VinSpinIn | | Descriptor: | 2-[4-[2-[[2-[3-[2-azanyl-5-(cyclopropylmethoxy)-3,3-dimethyl-indol-6-yl]oxypropyl]-1,3-dihydroisoindol-5-yl]oxy]ethyl]-1,2,3-triazol-1-yl]-1-[4-(2-pyrrolidin-1-ylethyl)piperidin-1-yl]ethanone, DIMETHYL SULFOXIDE, GLYCINE, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6KAF
 
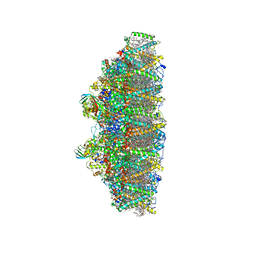 | | C2S2M2N2-type PSII-LHCII | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Chang, S.H, Shen, L.L, Huang, Z.H, Wang, W.D, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2019-06-22 | | Release date: | 2019-10-23 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure of a C2S2M2N2-type PSII-LHCII supercomplex from the green algaChlamydomonas reinhardtii.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KHJ
 
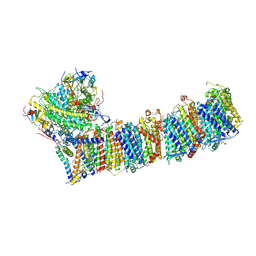 | | Supercomplex for electron transfer | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Pan, X, Cao, D, Xie, F, Zhang, X, Li, M. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase.
Nat Commun, 11, 2020
|
|
4QFG
 
 | |
6KMA
 
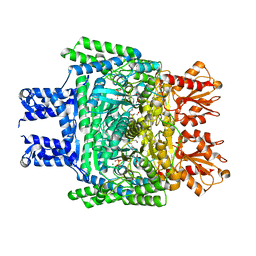 | | Crystal structure of SucA with glycolaldehyde-1-13C from Vibrio vulnificus | | Descriptor: | 2-oxidanylethanal, CALCIUM ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Seo, P.W, Kim, J.S. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Understanding the molecular properties of the E1 subunit (SucA) of alpha-ketoglutarate dehydrogenase complex from Vibrio vulnificus for the enantioselective ligation of acetaldehydes into (R)-acetoin.
Catalysis Science And Technology, 2020
|
|
3NNR
 
 | |
