4YWL
 
 | |
2X9A
 
 | | crystal structure of g3p from phage IF1 in complex with its coreceptor, the C-terminal domain of TolA | | Descriptor: | ATTACHMENT PROTEIN G3P, MEMBRANE SPANNING PROTEIN, REQUIRED FOR OUTER MEMBRANE INTEGRITY | | Authors: | Lorenz, S.H, Jakob, R.P, Dobbek, H, Schmid, F.X. | | Deposit date: | 2010-03-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The Filamentous Phages Fd and If1 Use Different Mechanisms to Infect Escherichia Coli.
J.Mol.Biol., 405, 2011
|
|
7AQK
 
 | | Model of the actin filament Arp2/3 complex branch junction in cells | | Descriptor: | Actin, alpha skeletal muscle, ACTA1, ... | | Authors: | Faessler, F, Dimchev, G, Hodirnau, V.V, Wan, W, Schur, F.K.M. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-electron tomography structure of Arp2/3 complex in cells reveals new insights into the branch junction.
Nat Commun, 11, 2020
|
|
7AST
 
 | | Apo Human RNA Polymerase III | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Ramsay, E.P, Abascal-Palacios, G, Daiss, J.L, King, H, Gouge, J, Pilsl, M, Beuron, F, Morris, E, Gunkel, P, Engel, C, Vannini, A. | | Deposit date: | 2020-10-28 | | Release date: | 2020-12-23 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of human RNA polymerase III.
Nat Commun, 11, 2020
|
|
6S68
 
 | | Structure of the Fluorescent Protein AausFP2 from Aequorea cf. australis at pH 7.6 | | Descriptor: | Aequorea cf. australis fluorescent protein 2 (AausFP2) | | Authors: | Depernet, H, Gotthard, G, Lambert, G.G, Shaner, N.C, Royant, A. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Aequorea's secrets revealed: New fluorescent proteins with unique properties for bioimaging and biosensing.
Plos Biol., 18, 2020
|
|
7AER
 
 | | Rebuilt and re-refined PDB entry 5yep: tri-AMPylated Shewanella oneidensis HEPN toxin in complex with MNT antitoxin | | Descriptor: | ADENOSINE MONOPHOSPHATE, Toxin-antitoxin system antidote Mnt family, Toxin-antitoxin system toxin HepN family | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-18 | | Release date: | 2020-12-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
7B2X
 
 | | Crystal structure of human 5' exonuclease Appollo H61Y variant | | Descriptor: | 5' exonuclease Apollo, NICKEL (II) ION | | Authors: | Newman, J.A, Baddock, H.T, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, von Delft, F, Arrowshmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-11-28 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A phosphate binding pocket is a key determinant of exo- versus endo-nucleolytic activity in the SNM1 nuclease family.
Nucleic Acids Res., 49, 2021
|
|
7X9F
 
 | | Crystal structure of actinomycin D-echinomycin-d(AGCGCGT/ACGCGCT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(P*AP*CP*GP*CP*GP*CP*T)-3'), DNA (5'-D(P*AP*GP*CP*GP*CP*GP*T)-3'), ... | | Authors: | Kao, S.H, Satange, R.B, Hou, M.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism.
Nucleic Acids Res., 50, 2022
|
|
7X6R
 
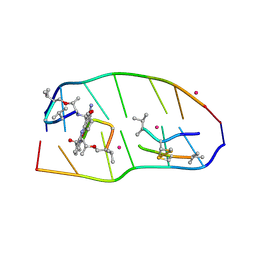 | | Crystal structure of actinomycin D-echinomycin-d(AGCACGT/ACGGGCT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, Actinomycin D, COBALT (II) ION, ... | | Authors: | Kao, S.H, Satange, R.B, Hou, M.H. | | Deposit date: | 2022-03-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism.
Nucleic Acids Res., 50, 2022
|
|
7X97
 
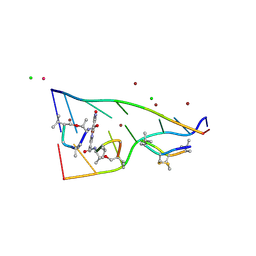 | | Crystal structure of actinomycin D-echinomycin-d(AGCCCGT/ACGGGCT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, Actinomycin D, CHLORIDE ION, ... | | Authors: | Kao, S.H, Satange, R.B, Hou, M.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism.
Nucleic Acids Res., 50, 2022
|
|
7XDJ
 
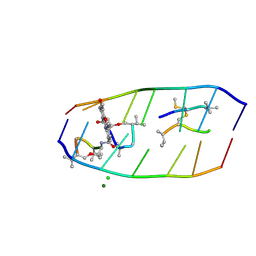 | | Crystal structure of actinomycin D-echinomycin-d(AGCGCGT/ACGAGCT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, CHLORIDE ION, DNA (5'-D(P*AP*CP*GP*AP*GP*CP*(BRU))-3'), ... | | Authors: | Chien, C.M, Satange, R.B, Hou, M.H. | | Deposit date: | 2022-03-27 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism.
Nucleic Acids Res., 50, 2022
|
|
7X1R
 
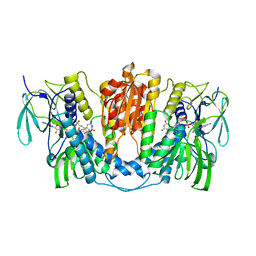 | | Cryo-EM structure of human thioredoxin reductase bound by Au | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, Thioredoxin reductase 1, ... | | Authors: | He, Z.S, Cao, P, Cao, S.H, He, B, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2022-02-24 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Au4 cluster inhibits human thioredoxin reductase activity via specifically binding of Au to Cys189
Nano Today, 47, 2022
|
|
5FKJ
 
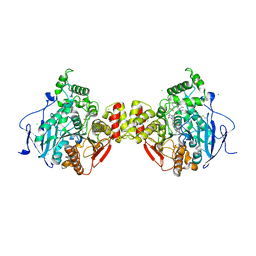 | | Crystal structure of mouse acetylcholinesterase in complex with C-547, an alkyl ammonium derivative of 6-methyl uracil | | Descriptor: | 1,3-BIS[5(DIETHYL-O-NITROBENZYLAMMONIUM)PENTYL]-6-METHYLURACIL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Nachon, F, Villard-Wandhammer, M, Petrov, K, Masson, P. | | Deposit date: | 2015-10-16 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.133 Å) | | Cite: | Slow-Binding Inhibition of Acetylcholinesterase by a 6-Methyluracil Alkyl-Ammonium Derivative: Mechanism and Advantages for Myasthenia Gravis Treatment.
Biochem.J., 473, 2016
|
|
4ZQQ
 
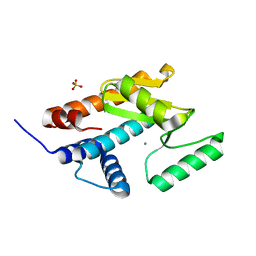 | | Apo form of influenza strain H1N1 polymerase acidic subunit N-terminal region | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein,Polymerase acidic protein, SULFATE ION | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Neya, S, Hoshino, T. | | Deposit date: | 2015-05-11 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit.
Biochemistry, 55, 2016
|
|
3O9R
 
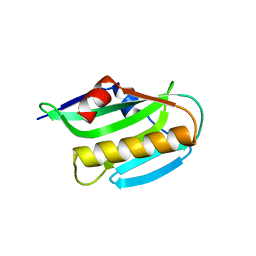 | | Effector domain of NS1 from influenza A/PR/8/34 containing a W187A mutation | | Descriptor: | Nonstructural protein 1 | | Authors: | Kerry, P.S, Lewis, A, Hale, B.G, Hass, C, Taylor, M.A, Randall, R.E, Russell, R.J.M. | | Deposit date: | 2010-08-04 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain.
Plos One, 6, 2011
|
|
1LBC
 
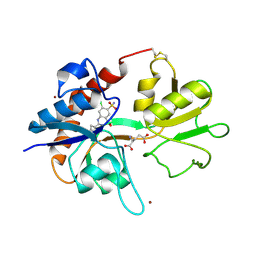 | | Crystal structure of GluR2 ligand binding core (S1S2J-N775S) in complex with cyclothiazide (CTZ) as well as glutamate at 1.8 A resolution | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamine Receptor 2, ... | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
1LBB
 
 | | Crystal structure of the GluR2 ligand binding domain mutant (S1S2J-N754D) in complex with kainate at 2.1 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamine receptor 2 | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
7WY4
 
 | | Structure of the CYP102A1 F87A Haem Domain with N-Enanthyl-L-Prolyl-L-Phenylalanine in complex with Styrene | | Descriptor: | (2S)-2-[[(2S)-1-heptylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, CHLORIDE ION, ... | | Authors: | Suzuki, K, Stanfield, J.K, Shisaka, Y, Omura, K, Kasai, C, Sugimoto, H, Shoji, O. | | Deposit date: | 2022-02-15 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Compound I Mimic Reveals the Transient Active Species of a Cytochrome P450 Enzyme: Insight into the Stereoselectivity of P450-Catalysed Oxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7WY3
 
 | | Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 F87V Mutant Haem Domain with N-(5-Cyclohexyl)valeroyl-L-Phenylalanine in complex with Styrene | | Descriptor: | (2~{S})-2-(5-cyclohexylpentanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, CHLORIDE ION, ... | | Authors: | Suzuki, K, Stanfield, J.K, Shisaka, Y, Omura, K, Kasai, C, Sugimoto, H, Shoji, O. | | Deposit date: | 2022-02-15 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Compound I Mimic Reveals the Transient Active Species of a Cytochrome P450 Enzyme: Insight into the Stereoselectivity of P450-Catalysed Oxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4ZI0
 
 | | Endonuclease inhibitor bound to influenza strain H1N1 polymerase acidic subunit N-terminal region without a chelation to the metal ions in the active site | | Descriptor: | 4-{(E)-[2-(4-chlorophenyl)hydrazinylidene]methyl}benzene-1,2,3-triol, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Neya, S, Hoshino, T. | | Deposit date: | 2015-04-27 | | Release date: | 2015-05-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit
Biochemistry, 55, 2016
|
|
7WY2
 
 | | Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 F87A Mutant Haem Domain with N-Enanthyl-L-Prolyl-L-Phenylalanine in complex with Styrene | | Descriptor: | (2S)-2-[[(2S)-1-heptylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, CHLORIDE ION, ... | | Authors: | Suzuki, K, Stanfield, J.K, Shisaka, Y, Omura, K, Kasai, C, Sugimoto, H, Shoji, O. | | Deposit date: | 2022-02-15 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Compound I Mimic Reveals the Transient Active Species of a Cytochrome P450 Enzyme: Insight into the Stereoselectivity of P450-Catalysed Oxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7WY1
 
 | | Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Enanthyl-L-Prolyl-L-Phenylalanine in complex with Styerene | | Descriptor: | (2S)-2-[[(2S)-1-heptylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, Oxomolybdenum Mesoporphyrin IX, ... | | Authors: | Suzuki, K, Stanfield, J.K, Shisaka, Y, Omura, K, Kasai, C, Sugimoto, H, Shoji, O. | | Deposit date: | 2022-02-15 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Compound I Mimic Reveals the Transient Active Species of a Cytochrome P450 Enzyme: Insight into the Stereoselectivity of P450-Catalysed Oxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5NL0
 
 | | Crystal structure of a 197-bp palindromic 601L nucleosome in complex with linker histone H1 | | Descriptor: | DNA (197-MER), Histone H1.0-B, Histone H2A type 1, ... | | Authors: | Garcia-Saez, I, Petosa, C, Dimitrov, S. | | Deposit date: | 2017-04-03 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (5.4 Å) | | Cite: | Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1.
Mol. Cell, 66, 2017
|
|
6RWK
 
 | |
5ZOK
 
 | | Crystal structure of human SMAD1-MAN1 complex. | | Descriptor: | Inner nuclear membrane protein Man1, Mothers against decapentaplegic homolog 1 | | Authors: | Miyazono, K, Ito, T, Tanokura, M. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis for receptor-regulated SMAD recognition by MAN1
Nucleic Acids Res., 46, 2018
|
|
