3ZL9
 
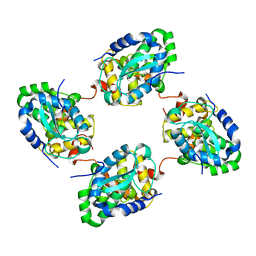 | | Crystal structure of the nucleocapsid protein from Schmallenberg virus | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
3F5M
 
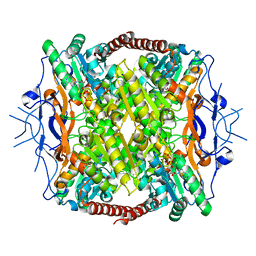 | | Crystal Structure of ATP-Bound Phosphofructokinase from Trypanosoma brucei | | Descriptor: | 6-phospho-1-fructokinase (ATP-dependent phosphofructokinase), ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | McNae, I.W, Martinez-Oyanedel, J, Keillor, J.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of ATP-bound phosphofructokinase from Trypanosoma brucei reveals conformational transitions different from those of other phosphofructokinases.
J.Mol.Biol., 385, 2009
|
|
4CQ4
 
 | | C-terminal fragment of Af1503-sol: transmembrane receptor Af1503 from Archaeoglobus fulgidus engineered for solubility | | Descriptor: | ENGINEERED VERSION OF TRANSMEMBRANE RECEPTOR AF1503 | | Authors: | Hartmann, M.D, Dunin-Horkawicz, S, Hulko, M, Martin, J, Coles, M, Lupas, A.N. | | Deposit date: | 2014-02-11 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Soluble Mutant of the Transmembrane Receptor Af1503 Features Strong Changes in Coiled-Coil Periodicity.
J.Struct.Biol., 186, 2014
|
|
4G63
 
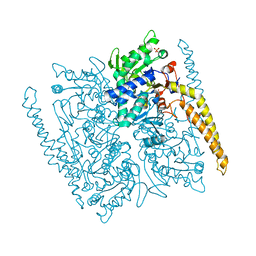 | | Crystal structure of cytosolic IMP-GMP specific 5'-nucleotidase (lpg0095) in complex with phosphate ions from Legionella pneumophila, Northeast Structural Genomics Consortium Target LgR1 | | Descriptor: | Cytosolic IMP-GMP specific 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Ho, C.K, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-07-18 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric regulation and substrate activation in cytosolic nucleotidase II from Legionella pneumophila.
Febs J., 281, 2014
|
|
3HEY
 
 | | Cyclic residues in alpha/beta-peptide helix bundles: GCN4-pLI side chain sequence on an (alpha-alpha-beta) backbone with cyclic beta-residues at positions 1, 4, 10, 19 and 28 | | Descriptor: | ACETATE ION, alpha/beta-peptide based on the GCN4-pLI side chain sequence with an (alpha-alpha-beta) backbone and cyclic beta-residues at positions 1, 4, ... | | Authors: | Horne, W.S, Price, J.L, Gellman, S.H. | | Deposit date: | 2009-05-10 | | Release date: | 2010-04-21 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural consequences of beta-amino acid preorganization in a self-assembling alpha/beta-peptide: fundamental studies of foldameric helix bundles.
J.Am.Chem.Soc., 132, 2010
|
|
3HEW
 
 | |
3HEU
 
 | |
3HET
 
 | |
3HEV
 
 | |
3HEX
 
 | | Cyclic residues in alpha/beta-peptide helix bundles: GCN4-pLI side chain sequence on an (alpha-alpha-beta) backbone with cyclic beta-residues at positions 1, 4, 19 and 28 | | Descriptor: | alpha/beta-peptide based on the GCN4-pLI side chain sequence with an (alpha-alpha-beta) backbone and cyclic beta-residues at positions 1, 4, 19 and 28 | | Authors: | Horne, W.S, Price, J.L, Gellman, S.H. | | Deposit date: | 2009-05-10 | | Release date: | 2010-04-21 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural consequences of beta-amino acid preorganization in a self-assembling alpha/beta-peptide: fundamental studies of foldameric helix bundles.
J.Am.Chem.Soc., 132, 2010
|
|
1MLD
 
 | | REFINED STRUCTURE OF MITOCHONDRIAL MALATE DEHYDROGENASE FROM PORCINE HEART AND THE CONSENSUS STRUCTURE FOR DICARBOXYLIC ACID OXIDOREDUCTASES | | Descriptor: | CITRIC ACID, MALATE DEHYDROGENASE | | Authors: | Gleason, W.B, Fu, Z, Birktoft, J.J, Banaszak, L.J. | | Deposit date: | 1994-01-24 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Refined crystal structure of mitochondrial malate dehydrogenase from porcine heart and the consensus structure for dicarboxylic acid oxidoreductases.
Biochemistry, 33, 1994
|
|
4W97
 
 | | Structure of ketosteroid transcriptional regulator KstR2 of Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, HTH-type transcriptional repressor KstR2, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] 3-[(3aS,4S,7aS)-7a-methyl-1,5-bis(oxidanylidene)-2,3,3a,4,6,7-hexahydroinden-4-yl]propanethioate | | Authors: | Stogios, P.J, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of a Ketosteroid Transcriptional Regulator of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
3CHF
 
 | |
3D1D
 
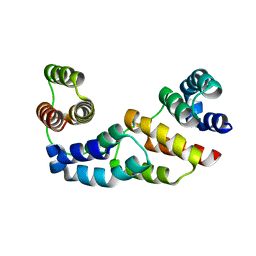 | | Hexagonal crystal structure of Tas3 C-terminal alpha motif | | Descriptor: | RNA-induced transcriptional silencing complex protein tas3 | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2008-05-05 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing.
Mol.Cell, 34, 2009
|
|
3ECA
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI L-ASPARAGINASE, AN ENZYME USED IN CANCER THERAPY (ELSPAR) | | Descriptor: | ASPARTIC ACID, L-asparaginase 2 | | Authors: | Swain, A.L, Jaskolski, M, Housset, D, Rao, J.K.M, Wlodawer, A. | | Deposit date: | 1993-07-02 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Escherichia coli L-asparaginase, an enzyme used in cancer therapy.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
4W94
 
 | | Crystal structure of cross-linked tetragonal hen egg white lysozyme soaked with 5mM [Ru(CO)3Cl2]2 | | Descriptor: | CHLORIDE ION, DIMETHYLFORMAMIDE, Lysozyme C, ... | | Authors: | Tabe, H, Fujita, K, Abe, S, Tsujimoto, M, Kuchimaru, T, Kizaka-Kondo, S, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Preparation of a Cross-Linked Porous Protein Crystal Containing Ru Carbonyl Complexes as a CO-Releasing Extracellular Scaffold
Inorg.Chem., 54, 2015
|
|
3D2O
 
 | | Crystal Structure of Manganese-metallated GTP Cyclohydrolase Type IB | | Descriptor: | AZIDE ION, CHLORIDE ION, LITHIUM ION, ... | | Authors: | Swairjo, M.A. | | Deposit date: | 2008-05-08 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Zinc-independent folate biosynthesis: genetic, biochemical, and structural investigations reveal new metal dependence for GTP cyclohydrolase IB
J.Bacteriol., 191, 2009
|
|
4MCT
 
 | |
4MCX
 
 | |
8IDS
 
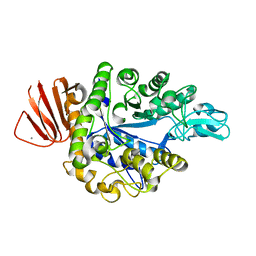 | | Crystal structure of Bacillus sp. AHU2216 GH13_31 Alpha-glucosidase E256Q/N258P in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Auiewiriyanukul, W, Saburi, W, Yu, J, Kato, K, Yao, M, Mori, H. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 alpha-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on beta → alpha Loop 5.
Molecules, 28, 2023
|
|
4LOR
 
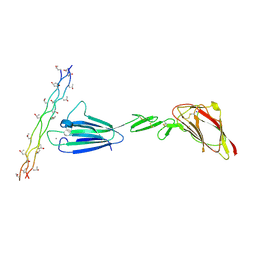 | | C1s CUB1-EGF-CUB2 in complex with a collagen-like peptide from C1q | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C1s subcomponent heavy chain, ... | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3CSC
 
 | |
3MQ6
 
 | |
4UUD
 
 | | Human dynamin 1 K44A superconstricted polymer stabilized with GTP | | Descriptor: | DYNAMIN-1 | | Authors: | Sundborger, A.C, Fang, S, Heymann, J.A, Ray, P, Chappie, J.S, Hinshaw, J.E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | A Dynamin Mutant Defines a Superconstricted Prefission State.
Cell Rep., 8, 2014
|
|
3CGL
 
 | |
