6Z69
 
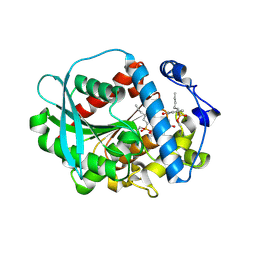 | | A novel metagenomic alpha/beta-fold esterase | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Acetyl esterase/lipase, MAGNESIUM ION, ... | | Authors: | Bollinger, A, Thies, S, Hoeppner, A, Kobus, S, Jaeger, K.-E, Smits, S.H.J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of a novel family IV esterase in free and substrate-bound form.
Febs J., 288, 2021
|
|
6G5L
 
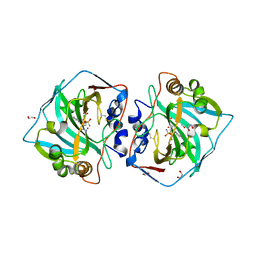 | | Crystal structure of human carbonic anhydrase isozyme XII with 4-chloro-2-(cyclohexylamino)-N-(2-hydroxyethyl)-5-sulfamoyl-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-(cyclohexylamino)-~{N}-(2-hydroxyethyl)-5-sulfamoyl-benzamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2018-03-29 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Design of two-tail compounds with rotationally fixed benzenesulfonamide ring as inhibitors of carbonic anhydrases.
Eur J Med Chem, 156, 2018
|
|
6G1E
 
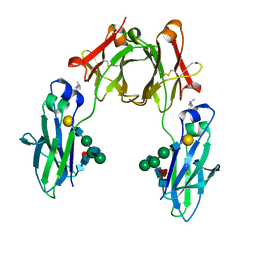 | | BEAT Fc with improved heterodimerization (Q3A-D84.4Q) | | Descriptor: | Immunoglobulin gamma-1 heavy chain, Immunoglobulin heavy constant gamma 1,Immunoglobulin heavy constant gamma 3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stutz, C, Blein, S. | | Deposit date: | 2018-03-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A single mutation increases heavy chain heterodimer assembly of bispecific antibodies by inducing structural disorder in one homodimer species.
J.Biol.Chem., 2020
|
|
5VT9
 
 | |
6G20
 
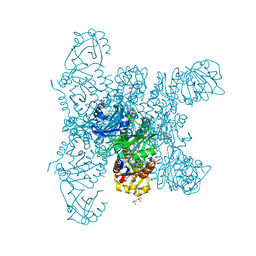 | | Crystal structure of a fluorescence optimized bathy phytochrome PAiRFP2 derived from wild-type Agp2 in its functional Meta-F intermediate state. | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Schmidt, A, Sauthof, L, Szczepek, M, Scheerer, P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural snapshot of a bacterial phytochrome in its functional intermediate state.
Nat Commun, 9, 2018
|
|
6ZGV
 
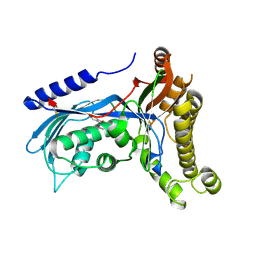 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, 2-(4-chlorophenyl)-~{N}-pyrimidin-2-yl-ethanamide, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
6Z51
 
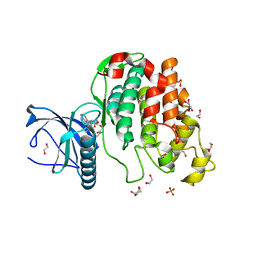 | | Crystal structure of CLK3 in complex with macrocycle ODS2002941 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3, N-cyclopentyl-2-[(11,15-dimethyl-10-oxo-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-6-yl)oxy]acetamide, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of CLK3 in complex with macrocycle ODS2002941
To Be Published
|
|
6Z5A
 
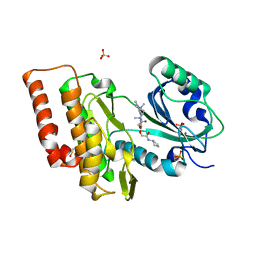 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2002941 | | Descriptor: | DIMETHYL SULFOXIDE, N-cyclopentyl-2-[(11,15-dimethyl-10-oxo-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-6-yl)oxy]acetamide, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2002941
To Be Published
|
|
6ZLN
 
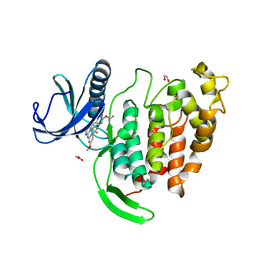 | | CLK1 bound with GW807982X (Cpd 8) | | Descriptor: | 1,2-ETHANEDIOL, 4-(6-ethoxypyrazolo[1,5-b]pyridazin-3-yl)-~{N}-[3-methoxy-5-(trifluoromethyl)phenyl]pyrimidin-2-amine, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6Z8V
 
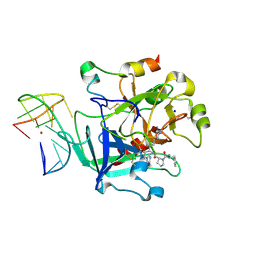 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3L), which contains 1-beta-D-lactopyranosyl residue in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
2AYH
 
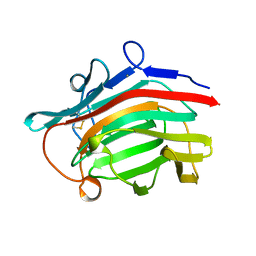 | | CRYSTAL AND MOLECULAR STRUCTURE AT 1.6 ANGSTROMS RESOLUTION OF THE HYBRID BACILLUS ENDO-1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE H(A16-M) | | Descriptor: | 1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Keitel, T, Heinemann, U. | | Deposit date: | 1995-02-02 | | Release date: | 1995-03-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal and molecular structure at 0.16-nm resolution of the hybrid Bacillus endo-1,3-1,4-beta-D-glucan 4-glucanohydrolase H(A16-M).
Eur.J.Biochem., 232, 1995
|
|
6G6U
 
 | | The dynamic nature of the VDAC1 channels in bilayers: human VDAC1 at 2.7 Angstrom resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, NITRATE ION, Voltage-dependent anion-selective channel protein 1 | | Authors: | Razeto, A, Gribbon, P, Loew, C. | | Deposit date: | 2018-04-03 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The dynamic nature of the VDAC1 channels in bilayers as revealed by two crystal structures of the human isoform in bicelles at 2.7 and 3.3 Angstrom resolution: implications for VDAC1 voltage-dependent mechanism and for its oligomerization
To Be Published
|
|
6Z2V
 
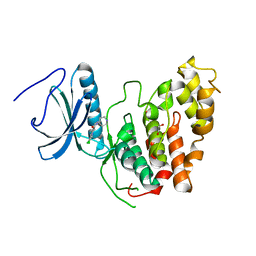 | | CLK3 A319V mutant bound with beta-carboline KH-CARB13 (Cpd 3) | | Descriptor: | (4~{S})-7,8-bis(chloranyl)-9-methyl-1-oxidanylidene-spiro[2,4-dihydropyrido[3,4-b]indole-3,4'-piperidine]-4-carbonitrile, 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3, ... | | Authors: | Schroeder, M, Chaikuad, A, Bracher, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
5EQZ
 
 | |
6Z4Y
 
 | | Crystal structure of Aurora A (STK6) in complex with macrocycle ODS2003208 | | Descriptor: | 1,2-ETHANEDIOL, 6-(2-methoxyethoxy)-11-methyl-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, Aurora kinase A, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of Aurora A (STK6) in complex with macrocycle ODS2003208
To Be Published
|
|
6Z56
 
 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2003208 | | Descriptor: | 6-(2-methoxyethoxy)-11-methyl-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, DIMETHYL SULFOXIDE, Serine/threonine-protein kinase haspin | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2003208
To Be Published
|
|
6Z59
 
 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2003816 | | Descriptor: | 11-oxa-8,14,18,19,22-pentazatetracyclo[13.5.2.12,6.018,21]tricosa-1(21),2,4,6(23),15(22),16,19-heptaen-7-one, SODIUM ION, Serine/threonine-protein kinase haspin | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2003816
To Be Published
|
|
6ZGM
 
 | | Crystal Structure of the VIM-2 Acquired Metallo-beta-Lactamase in Complex with the thiazolecarboxylate inhibitor ANT2681 | | Descriptor: | 5-[[4-(carbamimidamidocarbamoylamino)-3,5-bis(fluoranyl)phenyl]sulfonylamino]-1,3-thiazole-4-carboxylic acid, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Docquier, J.D, Pozzi, C, Marcoccia, F, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | ANT2681: SAR Studies Leading to the Identification of a Metallo-beta-lactamase Inhibitor with Potential for Clinical Use in Combination with Meropenem for the Treatment of Infections Caused by NDM-ProducingEnterobacteriaceae.
Acs Infect Dis., 6, 2020
|
|
6FNX
 
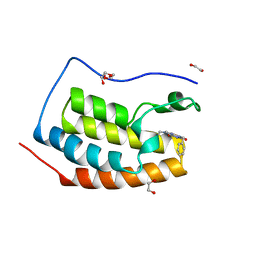 | | FIRST DOMAIN OF HUMAN BROMODOMAIN BRD4 IN COMPLEX WITH INHIBITOR F1 | | Descriptor: | 1,2-ETHANEDIOL, 7-ethyl-3-(phenylmethyl)purine-2,6-dione, Bromodomain-containing protein 4 | | Authors: | Raux, B, Betzi, S. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Integrated Strategy for Lead Optimization Based on Fragment Growing: The Diversity-Oriented-Target-Focused-Synthesis Approach.
J. Med. Chem., 61, 2018
|
|
6ZGW
 
 | | Structure of human galactokinase 1 bound with (4-chlorophenyl)methyl pyridine-3-carboxylate | | Descriptor: | (4-chlorophenyl)methyl pyridine-3-carboxylate, 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
6ZHD
 
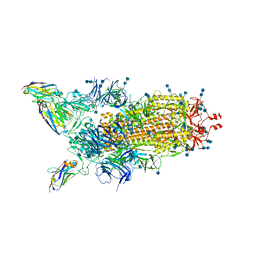 | | H11-H4 bound to Spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-H4, ... | | Authors: | Clare, D.K, Naismith, J.H, Weckener, M, Vogirala, V.K. | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | H11-H4 bound to Spike
To Be Published
|
|
6ZHK
 
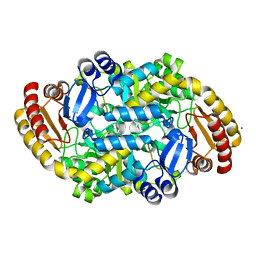 | | Crystal structure of adenosylmethionine-8-amino-7-oxononanoate aminotransferase from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, MAGNESIUM ION | | Authors: | Boyko, K.M, Nikolaeva, T.N, Stekhanova, T.N, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2020-06-23 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-Dimensional Structure of Thermostable D-Amino Acid Transaminase from the Archaeon Methanocaldococcus jannaschii DSM 2661
Crystallography Reports, 2021
|
|
6Z4C
 
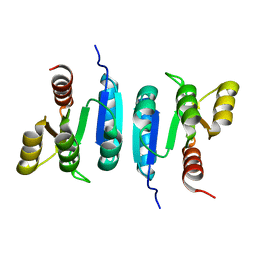 | | The structure of the N-terminal domain of RssB from E. coli | | Descriptor: | Regulator of RpoS | | Authors: | Zeth, K, Dimce, M, Terrence, D.M, Schuenemann, V, Dougan, D. | | Deposit date: | 2020-05-25 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into the RssB-Mediated Recognition and Delivery of sigma s to the AAA+ Protease, ClpXP.
Biomolecules, 10, 2020
|
|
6ZI3
 
 | | Crystal structure of OleP-6DEB bound to L-rhamnose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, ... | | Authors: | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Freda, I. | | Deposit date: | 2020-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
6Z52
 
 | | Crystal structure of CLK3 in complex with macrocycle ODS2003136 | | Descriptor: | 1,2-ETHANEDIOL, 11,15-dimethyl-6-(4-methylpiperazin-1-yl)-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of CLK3 in complex with macrocycle ODS2003136
To Be Published
|
|
