6T1W
 
 | |
6T24
 
 | | Cryo-EM structure of jasplakinolide-stabilized F-actin (aged) | | Descriptor: | (4~{R},7~{R},10~{S},13~{S},15~{E},19~{S})-10-(4-azanylbutyl)-4-(4-hydroxyphenyl)-7-(1~{H}-indol-3-ylmethyl)-8,13,15,19-tetramethyl-1-oxa-5,8,11-triazacyclononadec-15-ene-2,6,9,12-tetrone, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Pospich, S, Merino, F, Raunser, S. | | Deposit date: | 2019-10-07 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Effects and Functional Implications of Phalloidin and Jasplakinolide Binding to Actin Filaments.
Structure, 28, 2020
|
|
5ZJY
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, LYS-LYS-ARG-TYR-SER-ARG-2JN-GLN-LEU-LEU-2JN-PHE | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, B, Uhring, M. | | Deposit date: | 2018-03-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
5ZK5
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, LYS-ARG-TYR-SER-ARG-GLU-GLN-LEU-LEU-MK8-PHE-GLN-ARG-MK8 | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, F, Uhring, M. | | Deposit date: | 2018-03-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
5ZKC
 
 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with NMS | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-methyl scopolamine | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
6T31
 
 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, Streptavidin | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
3STL
 
 | | KcsA potassium channel mutant Y82C with Cadmium bound | | Descriptor: | CADMIUM ION, POTASSIUM ION, Voltage-gated potassium channel, ... | | Authors: | Raghuraman, H, Cordero-Morales, J, Jogini, V, Perozo, E. | | Deposit date: | 2011-07-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Cd(2+) Coordination during Slow Inactivation in Potassium Channels.
Structure, 20, 2012
|
|
3GB4
 
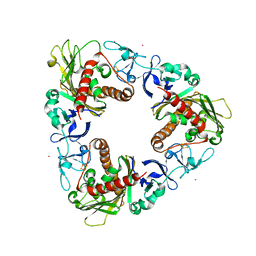 | | Crystal Structure of Dicamba Monooxygenase with Non-heme Cobalt and Dicamba | | Descriptor: | 3,6-dichloro-2-methoxybenzoic acid, COBALT (II) ION, DdmC, ... | | Authors: | Rydel, T.J, Sturman, E.J, Moshiri, F, Brown, G.R, Qi, Y. | | Deposit date: | 2009-02-18 | | Release date: | 2009-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dicamba monooxygenase: structural insights into a dynamic Rieske oxygenase that catalyzes an exocyclic monooxygenation.
J.Mol.Biol., 392, 2009
|
|
4DBL
 
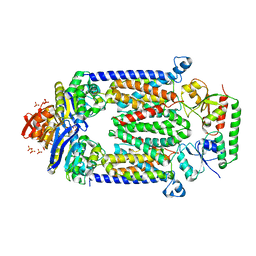 | | Crystal structure of E159Q mutant of BtuCDF | | Descriptor: | PHOSPHATE ION, SULFATE ION, Vitamin B12 import ATP-binding protein BtuD, ... | | Authors: | Korkhov, V.M, Mireku, S.M, Hvorup, R.N, Locher, K.P. | | Deposit date: | 2012-01-16 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.493 Å) | | Cite: | Asymmetric states of vitamin B12 transporter BtuCD are not discriminated by its cognate substrate binding protein BtuF.
Febs Lett., 586, 2012
|
|
5YWE
 
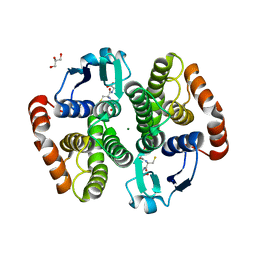 | | Crystal structure of hematopoietic prostaglandin D synthase apo form | | Descriptor: | GLUTATHIONE, GLYCEROL, Hematopoietic prostaglandin D synthase, ... | | Authors: | Kamo, M, Furubayashi, N, Inaka, K, Aritake, K, Urade, Y. | | Deposit date: | 2017-11-29 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of hematopoietic prostaglandin D synthase apo form
To Be Published
|
|
6T2U
 
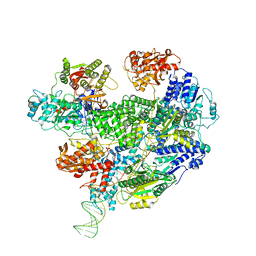 | | Cryo-EM structure of the RecBCD in complex with Chi-minus2 substrate | | Descriptor: | DNA (Chi-minus2), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-10-09 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6F0M
 
 | | GLIC mutant E35Q | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Full mutational mapping of titratable residues helps to identify proton-sensors involved in the control of channel gating in the Gloeobacter violaceus pentameric ligand-gated ion channel.
PLoS Biol., 15, 2017
|
|
3SVP
 
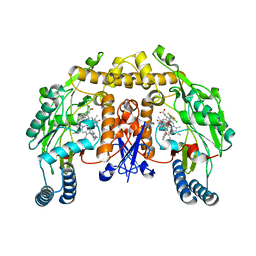 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(((3R,4R)-4-(2-((2,2-Difluoro-2-(3-chloro-5-fluorophenyl)ethyl)amino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[2-(3-chloro-5-fluorophenyl)-2,2-difluoroethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2011-07-12 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Improved Synthesis of Chiral Pyrrolidine Inhibitors and Their Binding Properties to Neuronal Nitric Oxide Synthase.
J.Med.Chem., 54, 2011
|
|
6F16
 
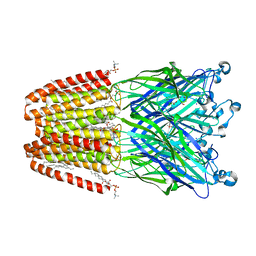 | | GLIC mutant H277Q | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Full mutational mapping of titratable residues helps to identify proton-sensors involved in the control of channel gating in the Gloeobacter violaceus pentameric ligand-gated ion channel.
PLoS Biol., 15, 2017
|
|
3KSO
 
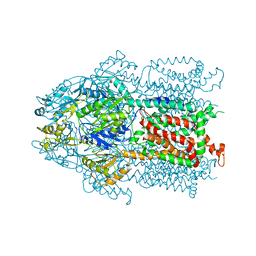 | | Structure and Mechanism of the Heavy Metal Transporter CusA | | Descriptor: | Cation efflux system protein cusA, SILVER ION | | Authors: | Su, C.-C. | | Deposit date: | 2009-11-23 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.367 Å) | | Cite: | Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport.
Nature, 467, 2010
|
|
3T6F
 
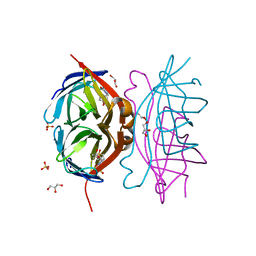 | | Biotin complex of Y54F core streptavidin | | Descriptor: | BIOTIN, BIOTIN-D-SULFOXIDE, GLYCEROL, ... | | Authors: | Baugh, L, Le Trong, I, Cerutti, D.S, Mehta, N, Gulich, S, Stayton, P.S, Stenkamp, R.E, Lybrand, T.P. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Second-Contact Shell Mutation Diminishes Streptavidin-Biotin Binding Affinity through Transmitted Effects on Equilibrium Dynamics.
Biochemistry, 51, 2012
|
|
3T6L
 
 | | Y54F mutant of core streptavidin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Streptavidin | | Authors: | Baugh, L, Le Trong, I, Stayton, P.S, Stenkamp, R.E, Lybrand, T.P. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Second-Contact Shell Mutation Diminishes Streptavidin-Biotin Binding Affinity through Transmitted Effects on Equilibrium Dynamics.
Biochemistry, 51, 2012
|
|
6TBU
 
 | | Structure of Drosophila melanogaster Dispatched | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Korkhov, V.M, Cannac, F. | | Deposit date: | 2019-11-04 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structure of the Hedgehog release protein Dispatched.
Sci Adv, 6, 2020
|
|
6T59
 
 | | Structure of rabbit 80S ribosome translating beta-tubulin in complex with tetratricopeptide protein 5 and nascent chain-associated complex | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Lin, Z, Gasic, I, Chandrasekaran, V, Peters, N, Shao, S, Ramakrishnan, V, Mitchison, T.J, Hegde, R.S. | | Deposit date: | 2019-10-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | TTC5 mediates autoregulation of tubulin via mRNA degradation.
Science, 367, 2020
|
|
6M49
 
 | | cryo-EM structure of Scap/Insig complex in the present of 25-hydroxyl cholesterol. | | Descriptor: | 25-HYDROXYCHOLESTEROL, Insulin-induced gene 2 protein, Sterol regulatory element-binding protein cleavage-activating protein,Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Yan, R, Cao, P, Song, W, Qian, H, Du, X, Coates, H.W, Zhao, X, Li, Y, Gao, S, Gong, X, Liu, X, Sui, J, Lei, J, Yang, H, Brown, A.J, Zhou, Q, Yan, C, Yan, N. | | Deposit date: | 2020-03-06 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols.
Science, 371, 2021
|
|
6EXY
 
 | | Neutron crystal structure of perdeuterated galectin-3C in complex with glycerol | | Descriptor: | GLYCEROL, Galectin-3 | | Authors: | Manzoni, F, Schrader, T.E, Ostermann, A, Oksanen, E, Logan, D.T. | | Deposit date: | 2017-11-10 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.1 Å), X-RAY DIFFRACTION | | Cite: | Elucidation of Hydrogen Bonding Patterns in Ligand-Free, Lactose- and Glycerol-Bound Galectin-3C by Neutron Crystallography to Guide Drug Design.
J. Med. Chem., 61, 2018
|
|
3FTU
 
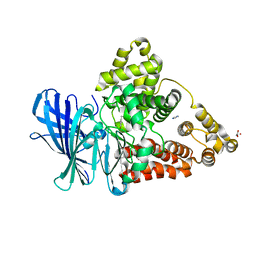 | | Leukotriene A4 hydrolase in complex with dihydroresveratrol | | Descriptor: | 5-[2-(4-hydroxyphenyl)ethyl]benzene-1,3-diol, ACETATE ION, IMIDAZOLE, ... | | Authors: | Davies, D.R. | | Deposit date: | 2009-01-13 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of leukotriene A4 hydrolase inhibitors using metabolomics biased fragment crystallography.
J.Med.Chem., 52, 2009
|
|
6TB2
 
 | |
6F4N
 
 | |
4BUR
 
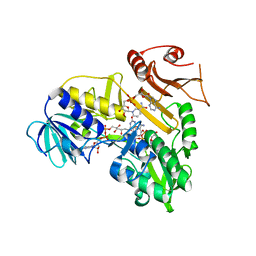 | | Crystal structure of the reduced human Apoptosis inducing factor complexed with NAD | | Descriptor: | APOPTOSIS INDUCING FACTOR 1, MITOCHONDRIAL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Martinez-Julvez, M, Herguedas, B, Hermoso, J.A, Ferreira, P, Villanueva, R, Medina, M. | | Deposit date: | 2013-06-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Insights Into the Coenzyme Mediated Monomer-Dimer Transition of the Pro-Apoptotic Apoptosis Inducing Factor.
Biochemistry, 53, 2014
|
|
