3OO9
 
 | |
3P2K
 
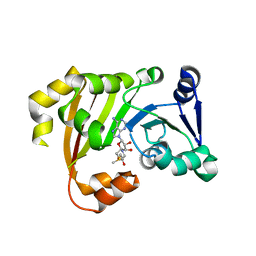 | | Structure of an antibiotic related Methyltransferase | | Descriptor: | 16S rRNA methylase, S-ADENOSYLMETHIONINE | | Authors: | Sivaraman, J, Husain, N. | | Deposit date: | 2010-10-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the methylation of A1408 in 16S rRNA by a panaminoglycoside resistance methyltransferase NpmA from a clinical isolate and analysis of the NpmA interactions with the 30S ribosomal subunit.
Nucleic Acids Res., 39, 2011
|
|
3OPO
 
 | |
3OQ1
 
 | | Crystal Structure of 11beta-Hydroxysteroid Dehydrogenase-1 (11b-HSD1) in Complex with Diarylsulfone Inhibitor | | Descriptor: | 3-(2-fluoroethyl)-4-({4-[(2S)-1,1,1-trifluoro-2-hydroxypropan-2-yl]phenyl}sulfonyl)benzonitrile, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2010-09-02 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The synthesis and SAR of novel diarylsulfone 11beta-HSD1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3P4E
 
 | | Phosphoribosylformylglycinamidine cyclo-ligase from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CITRIC ACID, ... | | Authors: | Osipiuk, J, Zhou, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-06 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Phosphoribosylformylglycinamidine cyclo-ligase from Vibrio cholerae.
To be Published
|
|
3OQJ
 
 | | Crystal structure of B. licheniformis CDPS yvmC-BLIC in complex with CAPSO | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, Putative uncharacterized protein yvmC | | Authors: | Bonnefond, L, Arai, T, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3P4U
 
 | | Crystal structure of active caspase-6 in complex with Ac-VEID-CHO inhibitor | | Descriptor: | Ac-VEID-CHO inhibitor, Caspase-6 | | Authors: | Mueller, I, Lamers, M.B.A.C, Ritchie, A.J, Dominguez, C, Munoz, I, Maillard, M, Kiselyov, A. | | Deposit date: | 2010-10-07 | | Release date: | 2011-09-28 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of active and inhibitor-bound human Casp6
To be Published
|
|
3OQS
 
 | | Crystal structure of importin-alpha bound to a CLIC4 NLS peptide | | Descriptor: | Importin subunit alpha-2, peptide of Chloride intracellular channel protein 4 | | Authors: | Mynott, A.V, Brown, L.J, Harrop, S.J, Curmi, P.M.G. | | Deposit date: | 2010-09-03 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of importin-alpha bound to a peptide bearing the nuclear localisation signal from chloride intracellular channel protein 4
Febs J., 278, 2011
|
|
3P5U
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3ORU
 
 | |
3OSD
 
 | |
3P6G
 
 | |
3P6S
 
 | |
3OT2
 
 | |
3P76
 
 | | X-ray crystal structure of Aquifex aeolicus LpxC complexed SCH1379777 | | Descriptor: | IMIDAZOLE, N-[(1S,2R)-2-hydroxy-1-(hydroxycarbamoyl)propyl]-4-[4-(phenylethynyl)phenyl]piperidine-1-carboxamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Orth, P. | | Deposit date: | 2010-10-12 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design and synthesis of potent Gram-negative specific LpxC inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3OTP
 
 | |
3P89
 
 | | FXR bound to a quinolinecarboxylic acid | | Descriptor: | 6-(4-{[3-(2,6-dichlorophenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}phenyl)quinoline-2-carboxylic acid, Farnesoid X receptor, Nuclear receptor coactivator 1, ... | | Authors: | Madauss, K.P, Williams, S.P, Deaton, D.N. | | Deposit date: | 2010-10-13 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformationally constrained farnesoid X receptor (FXR) agonists: Heteroaryl replacements of the naphthalene.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3P8F
 
 | |
3P92
 
 | |
3P9U
 
 | | Crystal structure of TetX2 from Bacteroides thetaiotaomicron with substrate analogue | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, TetX2 protein | | Authors: | Walkiewicz, K, Davlieva, M, Sun, C, Lau, K, Shamoo, Y. | | Deposit date: | 2010-10-18 | | Release date: | 2011-04-27 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of Bacteroides thetaiotaomicron TetX2: a tetracycline degrading monooxygenase at 2.8 A resolution.
Proteins, 79, 2011
|
|
3PA3
 
 | | X-ray crystal structure of compound 70 bound to human CHK1 kinase domain | | Descriptor: | 2-(4-chlorophenyl)-4-[(3S)-piperidin-3-ylamino]thieno[2,3-d]pyridazine-7-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, synthesis and SAR of thienopyridines as potent CHK1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OW2
 
 | | Crystal Structure of Enhanced Macrolide Bound to 50S Ribosomal Subunit | | Descriptor: | (2R,3S,4R,5R,8R,10R,11R,12S,13S,14R)-2-ethyl-3,4,10-trihydroxy-3,5,6,8,10,12,14-heptamethyl-15-oxo-11-[(3,4,6-trideoxy-3-{[3-(1-{(1S,2R)-1-(fluoromethyl)-2-hydroxy-2-[4-(methylsulfonyl)phenyl]ethyl}-1H-1,2,3-triazol-4-yl)propyl](methyl)amino}-beta-D-xylo-hexopyranosyl)oxy]-1-oxa-6-azacyclopentadecan-13-yl 2,6-dideoxy-3-C-methyl-3-O-methyl-alpha-L-ribo-hexopyranoside, 23S RIBOSOMAL RNA, 50S ribosomal protein L10E, ... | | Authors: | Kanyo, Z.F. | | Deposit date: | 2010-09-17 | | Release date: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: |
|
|
3OM5
 
 | | Crystal structure of B. megaterium levansucrase mutant N252A | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Strube, C.P, Homann, A, Gamer, M, Jahn, D, Seibel, J, Heinz, D.W. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Polysaccharide Synthesis of the Levansucrase SacB from Bacillus megaterium Is Controlled by Distinct Surface Motifs.
J.Biol.Chem., 286, 2011
|
|
3P39
 
 | |
3OMN
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with D132A mutation in the reduced state | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
