8T1C
 
 | | Cryo-EM structure of human TRPV4 ankyrin repeat domain in complex with GTPase RhoA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, Transient receptor potential cation channel subfamily V member 4-Enhanced green fluorescent protein chimera | | Authors: | Nadezhdin, K.D, Talyzina, I.A, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
8S47
 
 | | CdaA-D07 complex | | Descriptor: | CHLORIDE ION, Diadenylate cyclase, N-(pyridin-2-yl)-2-(1,3,5-trimethyl-1H-pyrazol-4-yl)acetamide | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2024-02-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | CdaA-D07 complex
To Be Published
|
|
8S4A
 
 | | CdaA-B04 complex | | Descriptor: | 2-[cyanomethyl(methyl)amino]-N-(6-methylpyridin-2-yl)ethanamide, CHLORIDE ION, Diadenylate cyclase | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2024-02-21 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | CdaA-B04 complex
To Be Published
|
|
1U4D
 
 | | Structure of the ACK1 Kinase Domain bound to Debromohymenialdisine | | Descriptor: | Activated CDC42 kinase 1, CHLORIDE ION, DEBROMOHYMENIALDISINE | | Authors: | Lougheed, J.C, Chen, R.H, Mak, P, Stout, T.J. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Phosphorylated and Unphosphorylated Kinase Domains of the Cdc42-associated Tyrosine Kinase ACK1.
J.Biol.Chem., 279, 2004
|
|
6YQD
 
 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) refined to 1.41 A in P212121 space group | | Descriptor: | Histidine triad nucleotide-binding protein 2, mitochondrial, POTASSIUM ION | | Authors: | Dolot, R.D, Wlodarczyk, A, Bujacz, G.D, Nawrot, B.C. | | Deposit date: | 2020-04-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.407 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6YI0
 
 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) refined to 1.65 A in P41212 space group | | Descriptor: | Histidine triad nucleotide-binding protein 2, mitochondrial, SODIUM ION | | Authors: | Dolot, R.D, Wlodarczyk, A, Bujacz, G.D, Nawrot, B.C. | | Deposit date: | 2020-03-31 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6YPX
 
 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) refined to 2.11 A in C2221 space group | | Descriptor: | CACODYLATE ION, GLYCEROL, Histidine triad nucleotide-binding protein 2, ... | | Authors: | Dolot, R.D, Wlodarczyk, A, Bujacz, G.D, Nawrot, B.C. | | Deposit date: | 2020-04-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6YPR
 
 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) refined to 1.26 A in H32 space group | | Descriptor: | GLYCEROL, Histidine triad nucleotide-binding protein 2, mitochondrial | | Authors: | Dolot, R.D, Wlodarczyk, A, Bujacz, G.D, Nawrot, B.C. | | Deposit date: | 2020-04-16 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6YJ8
 
 | | DarB-APO | | Descriptor: | ACETATE ION, CBS domain-containing protein YkuL, DI(HYDROXYETHYL)ETHER | | Authors: | Heidemann, J.L, Neumann, P, Ficner, R. | | Deposit date: | 2020-04-02 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | DarB from B. subtilis
To Be Published
|
|
6YVP
 
 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) complexed with dGMP and refined to 2.77 A | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, Histidine triad nucleotide-binding protein 2, mitochondrial | | Authors: | Dolot, R.D, Krakowiak, A, Nawrot, B.C. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6ZY3
 
 | | Cryo-EM structure of MlaFEDB in complex with phospholipid | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6AC1
 
 | | Rat Xanthine oxidoreductase, NADH bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Okamoto, K, Kawaguchi, Y. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Rat Xanthine oxidoreductase, D428A variant, NAD bound form
To Be Published
|
|
4XBA
 
 | | Hnt3 | | Descriptor: | Aprataxin-like protein, GLYCEROL, GUANOSINE, ... | | Authors: | Jacewicz, A, Chauleau, M, Shuman, S. | | Deposit date: | 2014-12-16 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | DNA3'pp5'G de-capping activity of aprataxin: effect of cap nucleoside analogs and structural basis for guanosine recognition.
Nucleic Acids Res., 43, 2015
|
|
6AC4
 
 | | Rat Xanthine oxidoreductase, D428N variant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Xanthine dehydrogenase/oxidase | | Authors: | Okamoto, K, Kawaguchi, Y. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Rat Xanthine oxidoreductase, D428A variant, NAD bound form
To Be Published
|
|
4XHF
 
 | | Crystal structure of Shewanella oneidensis NqrC | | Descriptor: | FLAVIN MONONUCLEOTIDE, Na-translocating NADH-quinone reductase subunit C NqrC, SODIUM ION | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2015-01-05 | | Release date: | 2015-12-16 | | Last modified: | 2016-03-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular insights into the enzymatic diversity of flavin-trafficking protein (Ftp; formerly ApbE) in flavoprotein biogenesis in the bacterial periplasm.
Microbiologyopen, 5, 2016
|
|
4XGV
 
 | | Crystal structure of Escherichia coli Flavin trafficking protein, an FMN transferase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | Molecular insights into the enzymatic diversity of flavin-trafficking protein (Ftp; formerly ApbE) in flavoprotein biogenesis in the bacterial periplasm.
Microbiologyopen, 5, 2016
|
|
4XDT
 
 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, a bifunctional FMN transferase/FAD pyrophosphatase, N55Y mutant, FAD bound form | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FAD:protein FMN transferase, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2014-12-19 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | Evidence for Posttranslational Protein Flavinylation in the Syphilis Spirochete Treponema pallidum: Structural and Biochemical Insights from the Catalytic Core of a Periplasmic Flavin-Trafficking Protein.
Mbio, 6, 2015
|
|
6ABU
 
 | | Rat Xanthine oxidoreductase, NAD bound form | | Descriptor: | BICARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Okamoto, K, Kawaguchi, Y. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Rat Xanthine oxidoreductase, NAD bound form
To Be Published
|
|
6AD4
 
 | | Rat Xanthine oxidoreductase, D428A variant, NADH bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Okamoto, K, Kawaguchi, Y. | | Deposit date: | 2018-07-30 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rat Xanthine oxidoreductase, D428A variant, NAD bound form
To Be Published
|
|
6AJU
 
 | | Rat Xanthine oxidoreductase | | Descriptor: | BICARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Okamoto, K, Kawaguchi, Y. | | Deposit date: | 2018-08-28 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Rat Xanthine oxidoreductase, D428A variant, NAD bound form
To Be Published
|
|
4XDU
 
 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein,a bifunctional FMN transferase/FAD pyrophosphatase, N55Y mutant, ADP bound form | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2014-12-20 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Evidence for Posttranslational Protein Flavinylation in the Syphilis Spirochete Treponema pallidum: Structural and Biochemical Insights from the Catalytic Core of a Periplasmic Flavin-Trafficking Protein.
Mbio, 6, 2015
|
|
4XYJ
 
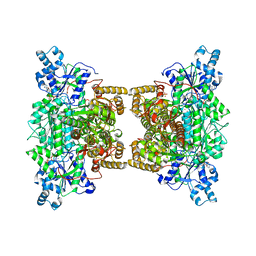 | | Crystal structure of human phosphofructokinase-1 in complex with ATP and Mg, Northeast Structural Genomics Consortium Target HR9275 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent 6-phosphofructokinase, platelet type, ... | | Authors: | Forouhar, F, Webb, B.A, Szu, F.-E, Seetharaman, J, Barber, D.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations.
Nature, 523, 2015
|
|
5IQ7
 
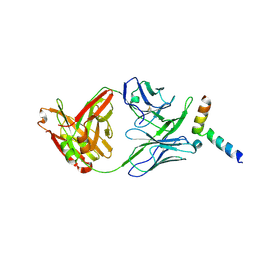 | | Crystal structure of 10E8-S74W Fab in complex with an HIV-1 gp41 peptide. | | Descriptor: | 10E8-S74W Heavy Chain, 10E8-S74W Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Kwon, Y.D, Caruso, W, Kwong, P.D. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2869 Å) | | Cite: | Optimization of the Solubility of HIV-1-Neutralizing Antibody 10E8 through Somatic Variation and Structure-Based Design.
J.Virol., 90, 2016
|
|
4XGX
 
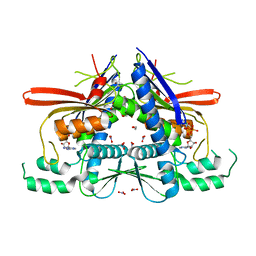 | | Crystal structure of Escherichia coli Flavin trafficking protein, an FMN transferase, Y60N mutant, ADP-inhibited | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, FAD:protein FMN transferase, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular insights into the enzymatic diversity of flavin-trafficking protein (Ftp; formerly ApbE) in flavoprotein biogenesis in the bacterial periplasm.
Microbiologyopen, 5, 2016
|
|
4XDR
 
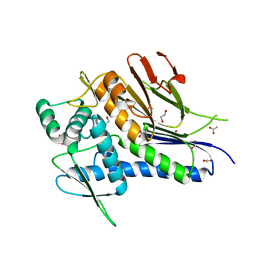 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, a bifunctional FMN transferase/FAD pyrophosphatase, D284A mutant, ADN bound form | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2014-12-19 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence for Posttranslational Protein Flavinylation in the Syphilis Spirochete Treponema pallidum: Structural and Biochemical Insights from the Catalytic Core of a Periplasmic Flavin-Trafficking Protein.
Mbio, 6, 2015
|
|
