5I55
 
 | | Crystal Structure of the Virulent PSM-alpha3 Peptide Forming a Cross-alpha amyloid-like Fibril | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Psm alpha-3 | | Authors: | Landau, M, Moshe, A, Tayeb-Fligelman, E, Sawaya, M.R, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2016-02-14 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The cytotoxic Staphylococcus aureus PSM alpha 3 reveals a cross-alpha amyloid-like fibril.
Science, 355, 2017
|
|
1EKY
 
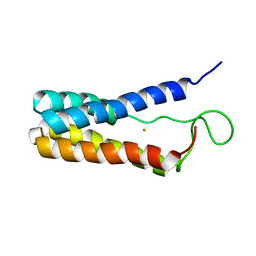 | |
1D7M
 
 | | COILED-COIL DIMERIZATION DOMAIN FROM CORTEXILLIN I | | Descriptor: | CORTEXILLIN I | | Authors: | Burkhard, P, Kammerer, R.A, Steinmetz, M.O, Bourenkov, G.P, Aebi, U. | | Deposit date: | 1999-10-19 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The coiled-coil trigger site of the rod domain of cortexillin I unveils a distinct network of interhelical and intrahelical salt bridges.
Structure Fold.Des., 8, 2000
|
|
1CIV
 
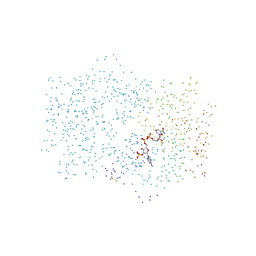 | | CHLOROPLAST NADP-DEPENDENT MALATE DEHYDROGENASE FROM FLAVERIA BIDENTIS | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-MALATE DEHYDROGENASE | | Authors: | Carr, P.D, Ashton, A.R, Verger, D, Ollis, D.L. | | Deposit date: | 1999-04-01 | | Release date: | 2000-04-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chloroplast NADP-malate dehydrogenase: structural basis of light-dependent regulation of activity by thiol oxidation and reduction.
Structure Fold.Des., 7, 1999
|
|
7SWQ
 
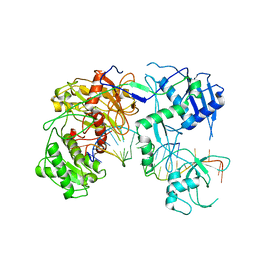 | |
5LC5
 
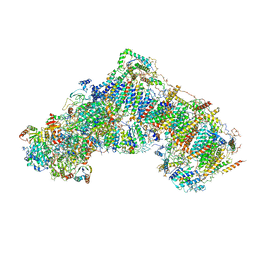 | | Structure of mammalian respiratory Complex I, class2 | | Descriptor: | Acyl carrier protein, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2016-06-19 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Structure of mammalian respiratory complex I.
Nature, 536, 2016
|
|
1CYA
 
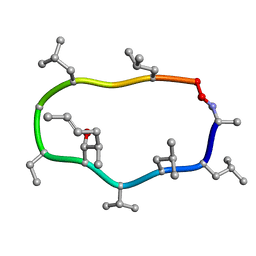 | |
7QXS
 
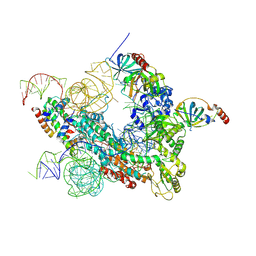 | | Cryo-EM structure of human telomerase-DNA-TPP1-POT1 complex (with POT1 side chains) | | Descriptor: | Adrenocortical dysplasia homolog (Mouse), isoform CRA_a, Histone H2A, ... | | Authors: | Sekne, Z, Ghanim, G.E, van Roon, A.M.M, Nguyen, T.H.D. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of human telomerase recruitment by TPP1-POT1.
Science, 375, 2022
|
|
1CJ2
 
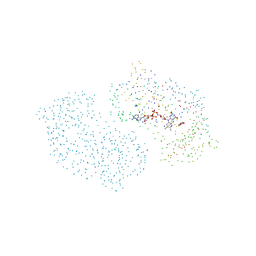 | | MUTANT GLN34ARG OF PARA-HYDROXYBENZOATE HYDROXYLASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Overkamp, K.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-04-21 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Switch of coenzyme specificity of p-hydroxybenzoate hydroxylase.
J.Mol.Biol., 292, 1999
|
|
7QXA
 
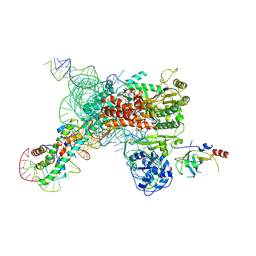 | | Cryo-EM map of human telomerase-DNA-TPP1 complex (sharpened) | | Descriptor: | Adrenocortical dysplasia homolog (Mouse), isoform CRA_a, Histone H2A, ... | | Authors: | Sekne, Z, Ghanim, G.E, van Roon, A.M.M, Nguyen, T.H.D. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-02 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of human telomerase recruitment by TPP1-POT1.
Science, 375, 2022
|
|
1CK2
 
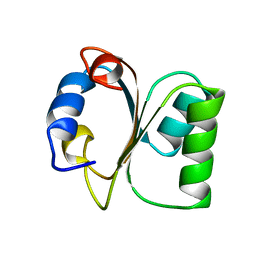 | |
7SVA
 
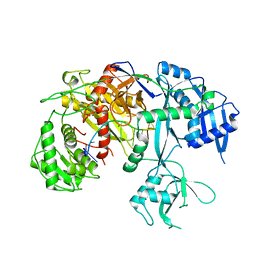 | | Cryo-EM structure of Arabidopsis Ago10-guide RNA complex | | Descriptor: | MAGNESIUM ION, Protein argonaute 10, RNA (5'-R(P*UP*GP*GP*AP*GP*UP*GP*UP*GP*AP*CP*AP*AP*UP*GP*GP*UP*GP*UP*UP*U)-3') | | Authors: | Xiao, Y, MacRae, I.J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for RNA slicing by a plant Argonaute.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7QXB
 
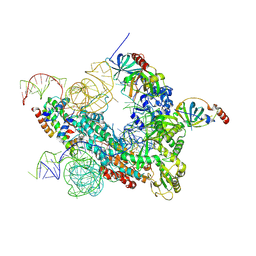 | | Cryo-EM map of human telomerase-DNA-TPP1-POT1 complex (sharpened map) | | Descriptor: | Adrenocortical dysplasia homolog (Mouse), isoform CRA_a, Histone H2A, ... | | Authors: | Sekne, Z, Ghanim, G.E, van Roon, A.M.M, Nguyen, T.H.D. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of human telomerase recruitment by TPP1-POT1.
Science, 375, 2022
|
|
1CVM
 
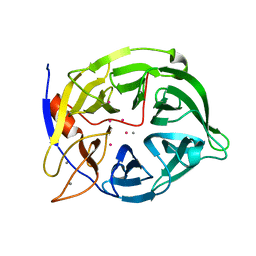 | | CADMIUM INHIBITED CRYSTAL STRUCTURE OF PHYTASE FROM BACILLUS AMYLOLIQUEFACIENS | | Descriptor: | CADMIUM ION, CALCIUM ION, PHYTASE | | Authors: | Shin, S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-08-24 | | Release date: | 2000-02-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
1KHP
 
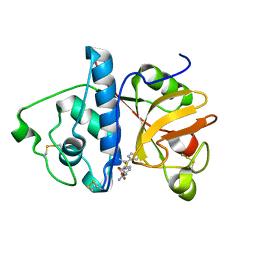 | | Monoclinic form of papain/ZLFG-DAM covalent complex | | Descriptor: | Papain, peptidic inhibitor | | Authors: | Janowski, R, Kozak, M, Jankowska, E, Grzonka, Z, Jaskolski, M. | | Deposit date: | 2001-11-30 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two polymorphs of a covalent complex between papain and a diazomethylketone inhibitor
J.Pept.Res., 64, 2004
|
|
1CL5
 
 | |
6ZKA
 
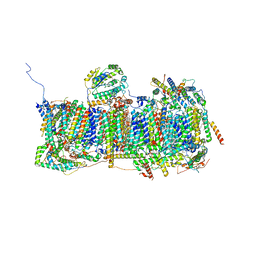 | | Membrane domain of open complex I during turnover | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6ZKP
 
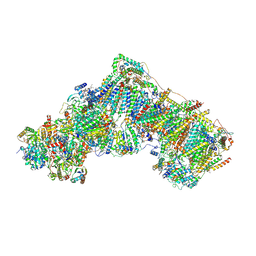 | | Native complex I, open1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
3RC2
 
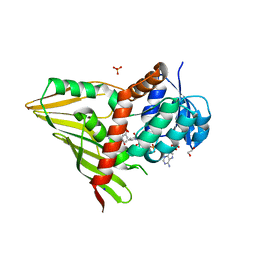 | | Crystal Structure of KijD10, a 3-ketoreductase from Actinomadura kijaniata in complex with TDP-benzene and NADP; open conformation | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phenoxy)phosphoryl]oxy}phosphoryl]thymidine, CHLORIDE ION, ... | | Authors: | Holden, H.M, Kubiak, R.L. | | Deposit date: | 2011-03-30 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined Structural and Functional Investigation of a C-3''-Ketoreductase Involved in the Biosynthesis of dTDP-l-Digitoxose.
Biochemistry, 50, 2011
|
|
1D7V
 
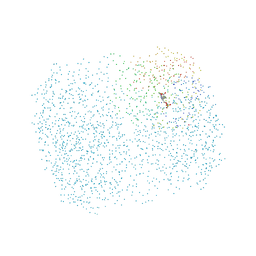 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH NMA | | Descriptor: | N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-2-METHYLALANINE, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
1CWP
 
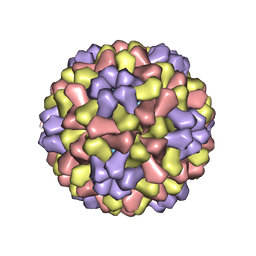 | | STRUCTURES OF THE NATIVE AND SWOLLEN FORMS OF COWPEA CHLOROTIC MOTTLE VIRUS DETERMINED BY X-RAY CRYSTALLOGRAPHY AND CRYO-ELECTRON MICROSCOPY | | Descriptor: | Coat protein, RNA (5'-R(*AP*U)-3'), RNA (5'-R(*AP*UP*AP*U)-3') | | Authors: | Speir, J.A, Johnson, J.E, Munshi, S, Wang, G, Timothy, S, Baker, T.S. | | Deposit date: | 1995-05-22 | | Release date: | 1995-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy.
Structure, 3, 1995
|
|
1D7U
 
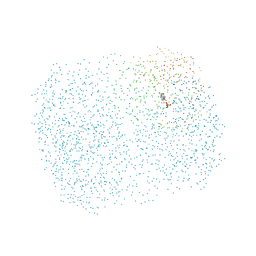 | | Crystal structure of the complex of 2,2-dialkylglycine decarboxylase with LCS | | Descriptor: | POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), SODIUM ION, ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
6ZKD
 
 | | Complex I during turnover, open1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6ZKT
 
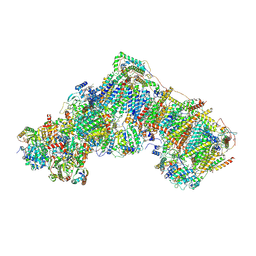 | | Deactive complex I, open2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
1D7R
 
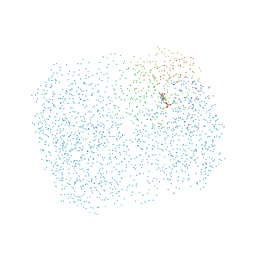 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH 5PA | | Descriptor: | N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
