4GKQ
 
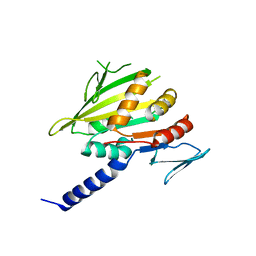 | |
1ZTD
 
 | | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus | | Descriptor: | Hypothetical Protein Pfu-631545-001 | | Authors: | Fu, Z.-Q, Horanyi, P, Florence, Q, Liu, Z.-J, Chen, L, Lee, D, Habel, J, Xu, H, Nguyen, D, Chang, S.-H, Zhou, W, Zhang, H, Jenney Jr, F.E, Sha, B, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-05-26 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus
To be Published
|
|
4FSR
 
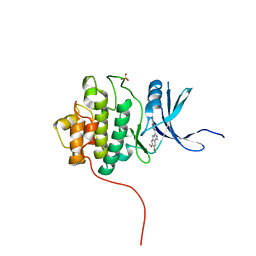 | | Crystal Structure of the CHK1 | | Descriptor: | 6,7-dimethoxy-3-[4-(1H-tetrazol-5-yl)phenyl]-1,4-dihydroindeno[1,2-c]pyrazole, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FV7
 
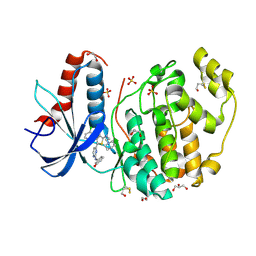 | | Crystal Structure of the ERK2 complexed with E94 | | Descriptor: | 1,2-ETHANEDIOL, 4-(3-chlorophenyl)-5-{2-[(3-hydroxyphenyl)amino]pyrimidin-4-yl}-2-{[2-(piperidin-1-yl)ethyl]amino}thiophene-3-carbonitrile, GLYCEROL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Xie, X. | | Deposit date: | 2012-06-29 | | Release date: | 2012-08-29 | | Last modified: | 2014-09-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the ERK2 complexed with E94
TO BE PUBLISHED
|
|
1ZX8
 
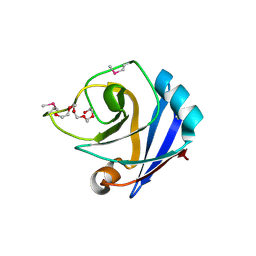 | |
4G2N
 
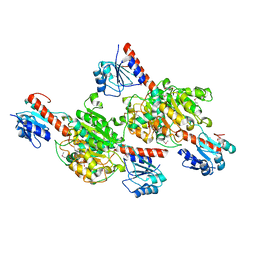 | | Crystal structure of putative D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding from Polaromonas sp. JS6 66 | | Descriptor: | CHLORIDE ION, D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-12 | | Release date: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding from Polaromonas sp. JS6 66
To be Published
|
|
4G3U
 
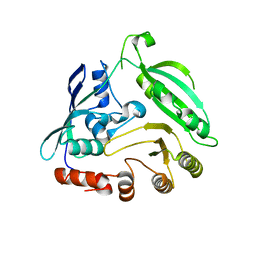 | |
4G5S
 
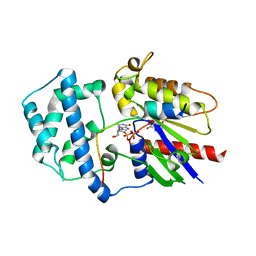 | | Structure of LGN GL3/Galphai3 complex | | Descriptor: | CITRIC ACID, G-protein-signaling modulator 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jia, M, Li, J, Zhu, J, Wen, W, Zhang, M, Wang, W. | | Deposit date: | 2012-07-18 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal Structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from Galphai subunits in G-coupled heterotrimeric proteins
To be Published
|
|
4G6N
 
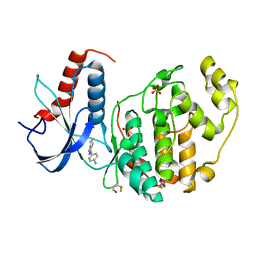 | | Crystal Structure of the ERK2 | | Descriptor: | 3-(4-chlorophenyl)-4,5,6,7-tetrahydro-1H-indazole, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Kang, Y.N, Stuckey, J.A, Xie, X. | | Deposit date: | 2012-07-19 | | Release date: | 2012-09-19 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the ERK2 complexed with EK0
to be published
|
|
1ZVH
 
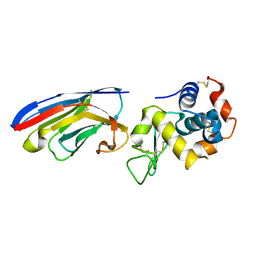 | | Crystal structure of the VHH domain D2-L24 in complex with hen egg white lysozyme | | Descriptor: | Immunoglobulin heavy chain antibody variable domain, Lysozyme C | | Authors: | De Genst, E, Silence, K, Decanniere, K, Conrath, K, Loris, R, Kinne, J, Muyldermans, S, Wyns, L. | | Deposit date: | 2005-06-02 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4GDM
 
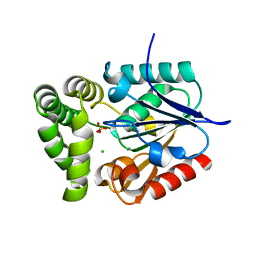 | | Crystal Structure of E.coli MenH | | Descriptor: | 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Johnston, J.M, Baker, E.N, Guo, Z, Jiang, M. | | Deposit date: | 2012-07-31 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of E. coli Native MenH and Two Active Site Mutants.
Plos One, 8, 2013
|
|
4GE4
 
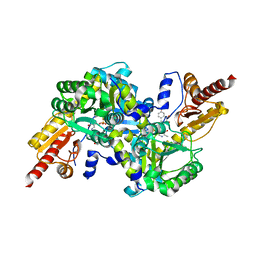 | | Kynurenine Aminotransferase II Inhibitors | | Descriptor: | (5-hydroxy-4-{[(1-hydroxy-7-methoxy-2-oxo-1,2-dihydroquinolin-3-yl)amino]methyl}-6-methylpyridin-3-yl)methyl dihydrogen phosphate, Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial | | Authors: | Pandit, J. | | Deposit date: | 2012-08-01 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-Based Design of Irreversible Human KAT II Inhibitors: Discovery of New Potency-Enhancing Interactions.
ACS Med Chem Lett, 4, 2013
|
|
3M97
 
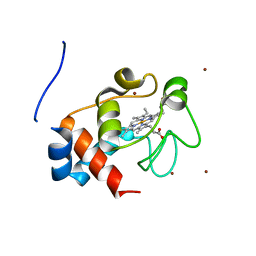 | | Structure of the soluble domain of cytochrome c552 with its flexible linker segment from Paracoccus denitrificans | | Descriptor: | Cytochrome c-552, HEME C, ZINC ION | | Authors: | Rajendran, C, Ermler, U, Ludwig, B, Michel, H. | | Deposit date: | 2010-03-20 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Structure at 1.5 A resolution of cytochrome c(552) with its flexible linker segment, a membrane-anchored protein from Paracoccus denitrificans.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1ZVF
 
 | | The crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from Saccharomyces cerevisiae | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, NICKEL (II) ION | | Authors: | Li, X, Guo, M, Teng, M, Niu, L. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from Saccharomyces cerevisiae
To be published
|
|
3M2U
 
 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | Deposit date: | 2010-03-08 | | Release date: | 2010-09-15 | | Last modified: | 2017-03-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
1ZYB
 
 | |
4FTE
 
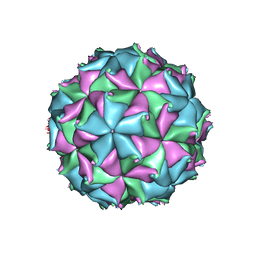 | | Crystal structure of the D75N mutant capsid of Flock House virus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
4FU7
 
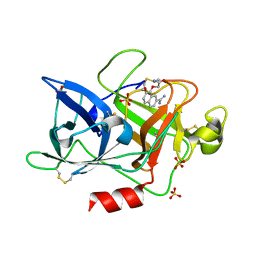 | | Crystal Structure of the Urokinase | | Descriptor: | 2-[(7-carbamimidoyl-2-methoxynaphthalen-1-yl)oxy]acetamide, ACETATE ION, SULFATE ION, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Nienaber, V, Giranda, V. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Urokinase
to be published
|
|
4FUO
 
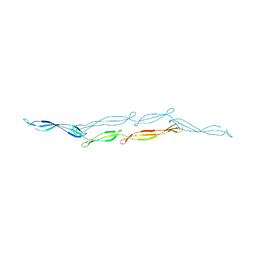 | |
4FV1
 
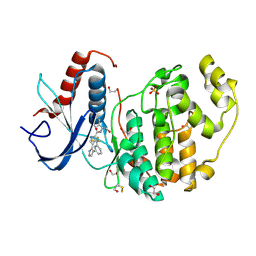 | |
4FW3
 
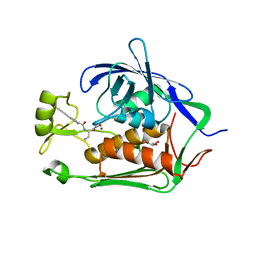 | |
4FSU
 
 | | Crystal Structure of the CHK1 | | Descriptor: | 4-[6-(1H-imidazol-1-ylmethyl)-7-methoxy-2,4-dihydroindeno[1,2-c]pyrazol-3-yl]benzonitrile, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4FT5
 
 | | Crystal Structure of the CHK1 | | Descriptor: | 1-{5-chloro-2-[(3R)-pyrrolidin-3-yloxy]phenyl}-3-(5-cyanopyrazin-2-yl)urea, GLYCEROL, SULFATE ION, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
4IBB
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | Polymerase cofactor VP35, {4-[(5R)-3-hydroxy-2-oxo-4-(thiophen-2-ylcarbonyl)-5-(2,4,5-trimethylphenyl)-2,5-dihydro-1H-pyrrol-1-yl]phenyl}acetic acid | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-08 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
1XMT
 
 | | X-ray structure of gene product from arabidopsis thaliana at1g77540 | | Descriptor: | BROMIDE ION, putative acetyltransferase | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
