5VHB
 
 | |
5VIB
 
 | |
5VI9
 
 | |
6ZEA
 
 | | Strictosidine Synthase from Catharanthus roseus in complex with racemic 1-methyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{R})-1-methyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, (1~{S})-1-methyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, 1,2-ETHANEDIOL, ... | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Strictosidine Synthase from Catharanthus roseus in complex with racemic 1-methyl-2,3,4,9-tetrahydro-1H-beta-carboline
To Be Published
|
|
2GH4
 
 | | YteR/D143N/dGalA-Rha | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose, Putative glycosyl hydrolase yteR | | Authors: | Itoh, T, Ochiai, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2006-03-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of unsaturated rhamnogalacturonyl hydrolase complexed with substrate
Biochem.Biophys.Res.Commun., 347, 2006
|
|
3HC7
 
 | |
2GEX
 
 | |
2GHB
 
 | | Thermotoga maritima maltotriose binding protein, ligand free form | | Descriptor: | maltose ABC transporter, periplasmic maltose-binding protein | | Authors: | Cuneo, M.J, Changela, A, Hocker, B, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2006-03-27 | | Release date: | 2007-02-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | T. maritima maltotriose binding protein open form
To be Published
|
|
7QZN
 
 | | Amine Dehydrogenase from Cystobacter fuscus (CfusAmDH) W145A mutant with NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amine Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Bennett, M, Ducrot, L, Vaxelaire-Vergne, C, Grogan, G. | | Deposit date: | 2022-01-31 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Cover Feature: Expanding the Substrate Scope of Native Amine Dehydrogenases through In Silico Structural Exploration and Targeted Protein Engineering (ChemCatChem 22/2022)
Chemcatchem, 14, 2022
|
|
7QZL
 
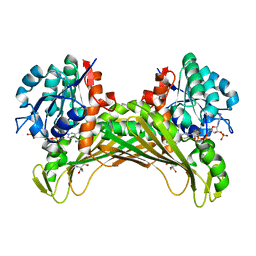 | | Amine Dehydrogenase from Cystobacter fuscus (CfusAmDH) W145A mutant with NADP+ and pentylamine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AMYLAMINE, Amine Dehydrogenase, ... | | Authors: | Bennett, M, Ducrot, L, Vaxelaire-Vergne, C, Grogan, G. | | Deposit date: | 2022-01-31 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cover Feature: Expanding the Substrate Scope of Native Amine Dehydrogenases through In Silico Structural Exploration and Targeted Protein Engineering (ChemCatChem 22/2022)
Chemcatchem, 14, 2022
|
|
7LII
 
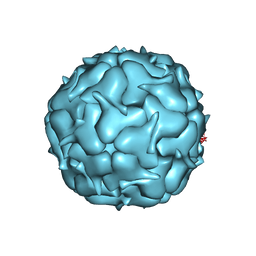 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S7D | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
7LIL
 
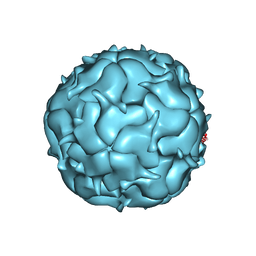 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant SE3 | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
7LIJ
 
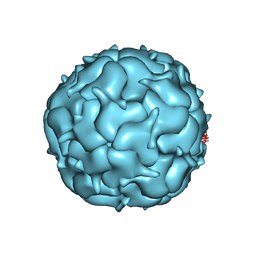 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S1K | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
7LIK
 
 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S1R | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
5KDD
 
 | | Apo-structure of humanised RadA-mutant humRadA22 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
6JYZ
 
 | | Crystal structure of endogalactoceramidase | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ISOPROPYL ALCOHOL, ... | | Authors: | Liuqing, C, Yan, F. | | Deposit date: | 2019-04-29 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of an endogalactosylceramidase from Rhodococcus hoagii 103S reveals the molecular basis of its substrate specificity.
J.Struct.Biol., 208, 2019
|
|
4F8V
 
 | | Crystal structure of the bacterial ribosomal decoding site in complex with sisomicin (P21212 form) | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(P*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Koganei, M, Kasahara, T. | | Deposit date: | 2012-05-18 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and specific binding mode of sisomicin to the bacterial ribosomal decoding site.
Acs Med.Chem.Lett., 3, 2012
|
|
6D69
 
 | |
7LIM
 
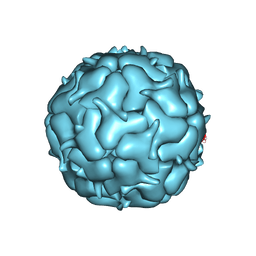 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S6E | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
7LIS
 
 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S5D | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
7LIT
 
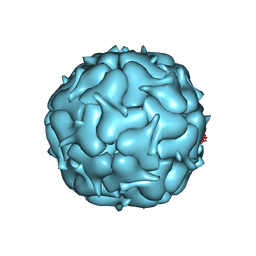 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S7G | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
5VRA
 
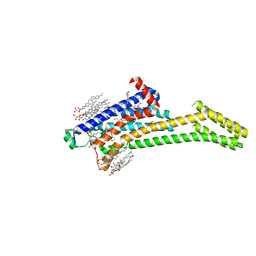 | | 2.35-Angstrom In situ Mylar structure of human A2A adenosine receptor at 100 K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Broecker, J, Morizumi, T, Ou, W.-L, Klingel, V, Kuo, A, Kissick, D.J, Ishchenko, A, Lee, M.-Y, Xu, S, Makarov, O, Cherezov, V, Ogata, C.M, Ernst, O.P. | | Deposit date: | 2017-05-10 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions.
Nat Protoc, 13, 2018
|
|
3HHX
 
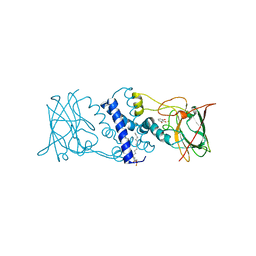 | | Crystal structure determination of Catechol 1,2-Dioxygenase from Rhodococcus opacus 1CP in complex with pyrogallol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, BENZENE-1,2,3-TRIOL, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Briganti, F. | | Deposit date: | 2009-05-18 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
5VZL
 
 | | cryo-EM structure of the Cas9-sgRNA-AcrIIA4 anti-CRISPR complex | | Descriptor: | CRISPR-associated endonuclease Cas9, phage anti-CRISPR AcrIIA4, single guide RNA (116-MER) | | Authors: | Jiang, F, Liu, J.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Disabling Cas9 by an anti-CRISPR DNA mimic.
Sci Adv, 3, 2017
|
|
1MML
 
 | | MECHANISTIC IMPLICATIONS FROM THE STRUCTURE OF A CATALYTIC FRAGMENT OF MMLV REVERSE TRANSCRIPTASE | | Descriptor: | MMLV REVERSE TRANSCRIPTASE | | Authors: | Georgiadis, M.M, Jessen, S.M, Ogata, C.M, Telesnitsky, A, Goff, S.P, Hendrickson, W.A. | | Deposit date: | 1995-07-18 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic implications from the structure of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase.
Structure, 3, 1995
|
|
