4AFJ
 
 | | 5-aryl-4-carboxamide-1,3-oxazoles: potent and selective GSK-3 inhibitors | | Descriptor: | 5-(4-METHOXYPHENYL)-N-(PYRIDIN-4-YLMETHYL)-1,3-OXAZOLE-4-CARBOXAMIDE, GLYCEROL, GLYCOGEN SYNTHASE KINASE-3 BETA, ... | | Authors: | Gentile, G, Merlo, G, Pozzan, A, Bernasconi, G, Bax, B, Bamborough, P, Bridges, A, Carter, P, Neu, M, Yao, G, Brough, C, Cutler, G, Coffin, A, Belyanskaya, S. | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | 5-Aryl-4-Carboxamide-1,3-Oxazoles: Potent and Selective Gsk-3 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1G08
 
 | | CARBONMONOXY LIGANDED BOVINE HEMOGLOBIN PH 5.0 | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN ALPHA CHAIN, HEMOGLOBIN BETA CHAIN, ... | | Authors: | Mueser, T.C, Rogers, P.H, Arnone, A. | | Deposit date: | 2000-10-05 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interface sliding as illustrated by the multiple quaternary structures of liganded hemoglobin.
Biochemistry, 39, 2000
|
|
8OW4
 
 | | 2.75 angstrom crystal structure of human NFAT1 with bound DNA | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*AP*TP*TP*TP*TP*TP*CP*CP*AP*GP*C)-3'), DNA (5'-D(*TP*TP*GP*CP*TP*GP*GP*AP*AP*AP*AP*AP*TP*AP*G)-3'), Nuclear factor of activated T-cells, ... | | Authors: | Lopez-Sagaseta, J, Erausquin, E, Hernandez-Morales, S, Urdiciain, A, Lasarte, J.J, Lozano, T. | | Deposit date: | 2023-04-27 | | Release date: | 2023-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 angstrom crystal structure of human NFAT1 with bound DNA
Not published
|
|
6FWL
 
 | | Structure of an E333Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex alpha-Glc-1,3-(1,2-anhydro-carba-mannose) | | Descriptor: | (1~{S},2~{S},3~{R},4~{R},6~{R})-4-(hydroxymethyl)-7-oxabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
6FWP
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-1,3-mannobiose and alpha-1,2-mannobiose | | Descriptor: | ACETATE ION, Glycosyl hydrolase family 71, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, ... | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
5Z2K
 
 | | Structure of S38A mutant Mn-bound periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Saini, G, Sharma, N, Dalal, V, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-01-02 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The analysis of subtle internal communications through mutation studies in periplasmic metal uptake protein CLas-ZnuA2
J. Struct. Biol., 204, 2018
|
|
6TNB
 
 | | X-RAY STRUCTURE OF MPS1 IN COMPLEX WITH COMPOUND 41 | | Descriptor: | (2~{R})-2-(4-fluorophenyl)-~{N}-[4-[2-[(2-methoxy-4-methylsulfonyl-phenyl)amino]-[1,2,4]triazolo[1,5-a]pyridin-6-yl]phenyl]propanamide, CHLORIDE ION, Dual specificity protein kinase TTK | | Authors: | Holton, S.J, Schulze, V.K, Klar, U, Kosemund, D, Siemeister, G, Bader, B, Prechtl, S, Briem, H, Marquardt, T, Schirok, H, Bohlmann, R, Nguyen, D, Fernandez-Montalvan, A, Boemer, U, Eberspaecher, U, Brands, M, Nussbaum, F, Koppitz, M. | | Deposit date: | 2019-12-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Treating Cancer by Spindle Assembly Checkpoint Abrogation: Discovery of Two Clinical Candidates, BAY 1161909 and BAY 1217389, Targeting MPS1 Kinase.
J.Med.Chem., 63, 2020
|
|
3ZRM
 
 | | Identification of 2-(4-pyridyl)thienopyridinones as GSK-3beta inhibitors | | Descriptor: | 7-(4-HYDROXYPHENYL)-2-PYRIDIN-4-YL-5H-THIENO[3,2-C]PYRIDIN-4-ONE, GLYCEROL, GLYCOGEN SYNTHASE KINASE-3 BETA, ... | | Authors: | Gentile, G, Bernasconi, G, Pozzan, A, Merlo, G, Marzorati, P, Bamborough, P, Bax, B, Bridges, A, Brough, C, Carter, P, Cutler, G, Neu, M, Takada, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Identification of 2-(4-Pyridyl)Thienopyridinones as Gsk-3Beta Inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6FWI
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-(1,2-anhydro-carba-mannosamine) | | Descriptor: | (1~{R},2~{R},3~{R},4~{R},6~{R})-4-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
3ZRK
 
 | | Identification of 2-(4-pyridyl)thienopyridinones as GSK-3beta inhibitors | | Descriptor: | 2-(4-PYRIDINYL)FURO[3,2-C]PYRIDIN-4(5H)-ONE, GLYCEROL, GLYCOGEN SYNTHASE KINASE-3 BETA, ... | | Authors: | Gentile, G, Bernasconi, G, Pozzan, A, Merlo, G, Marzorati, P, Bamborough, P, Bax, B, Bridges, A, Brough, C, Carter, P, Cutler, G, Neu, M, Takada, M. | | Deposit date: | 2011-06-16 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Identification of 2-(4-Pyridyl)Thienopyridinones as Gsk-3Beta Inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5Z2J
 
 | | structure of S38A mutant metal-free periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, GLYCEROL, Periplasmic solute binding protein, ... | | Authors: | Saini, G, Sharma, N, Dalal, V, Kumar, P, Sharma, A.K. | | Deposit date: | 2018-01-02 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The analysis of subtle internal communications through mutation studies in periplasmic metal uptake protein CLas-ZnuA2
J. Struct. Biol., 204, 2018
|
|
3ZRL
 
 | | Identification of 2-(4-pyridyl)thienopyridinones as GSK-3beta inhibitors | | Descriptor: | 7-BROMO-2-PYRIDIN-4-YL-5H-THIENO[3,2-C]PYRIDIN-4-ONE, GLYCEROL, GLYCOGEN SYNTHASE KINASE-3 BETA, ... | | Authors: | Gentile, G, Bernasconi, G, Pozzan, A, Merlo, G, Marzorati, P, Bamborough, P, Bax, B, Bridges, A, Brough, C, Carter, P, Cutler, G, Neu, M, Takada, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Identification of 2-(4-Pyridyl)Thienopyridinones as Gsk-3Beta Inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6FWO
 
 | | Structure of an E336Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-1,2-anhydro-mannose hydrolyzed by enzyme | | Descriptor: | (1~{S},2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
6WU8
 
 | | Structure of human SHP2 in complex with inhibitor IACS-13909 | | Descriptor: | 1-[3-(2,3-dichlorophenyl)-1H-pyrazolo[3,4-b]pyrazin-6-yl]-4-methylpiperidin-4-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Leonard, P.G, Joseph, S, Rodenberger, A. | | Deposit date: | 2020-05-04 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric SHP2 Inhibitor, IACS-13909, Overcomes EGFR-Dependent and EGFR-Independent Resistance Mechanisms toward Osimertinib.
Cancer Res., 80, 2020
|
|
2H0W
 
 | |
5LFG
 
 | | X-ray structure of a new fully ligated carbomonoxy form of Trematomus newnesi hemoglobin (Hb1TnCO). | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha-1, Hemoglobin subunit beta-1/2, ... | | Authors: | Vitagliano, L, Mazzarella, L, Merlino, A, Vergara, A. | | Deposit date: | 2016-07-01 | | Release date: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Fine Sampling of the RT Quaternary-Structure Transition of a Tetrameric Hemoglobin.
Chemistry, 23, 2017
|
|
8OUO
 
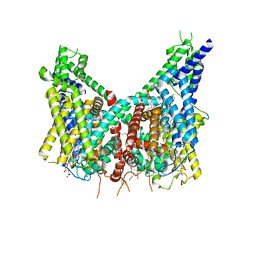 | | Human TPC2 in Complex with Antagonist (S)-SG-094 | | Descriptor: | (1S)-6-methoxy-2-methyl-7-phenoxy-1-[(4-phenoxyphenyl)methyl]-3,4-dihydro-1H-isoquinoline, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Chi, G, Pike, A.C.W, Maclean, E.M, Li, H, Mukhopadhyay, S.M.M, Bohstedt, T, Wang, D, McKinley, G, Fernandez-Cid, A, Duerr, K. | | Deposit date: | 2023-04-24 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for inhibition of the lysosomal two-pore channel TPC2 by a small molecule antagonist.
Structure, 2024
|
|
8P4R
 
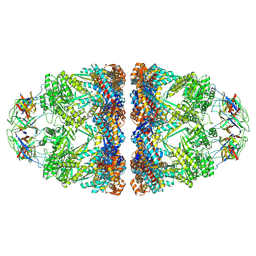 | | In situ structure average of GroEL14-GroES14 complexes in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (11.9 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography
Nature, 2024
|
|
8P4P
 
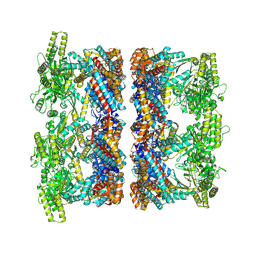 | | Structure average of GroEL14 complexes found in the cytosol of Escherichia coli overexpressing GroEL obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography
Nature, 2024
|
|
8P4O
 
 | | CryoEM structure of a GroEL7-GroES7 cage with encapsulated ordered substrate MetK in the presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Beck, F, Bracher, A, Caravajal, A.I, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography
Nature, 2024
|
|
2Z2U
 
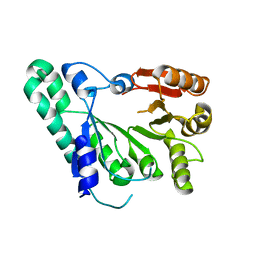 | | Crystal structure of archaeal TYW1 | | Descriptor: | UPF0026 protein MJ0257 | | Authors: | Suzuki, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2007-05-28 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Radical SAM Enzyme Catalyzing Tricyclic Modified Base Formation in tRNA
J.Mol.Biol., 372, 2007
|
|
5OQ0
 
 | |
3NF5
 
 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
3TGZ
 
 | |
2DEF
 
 | |
