2N8S
 
 | |
1XTM
 
 | | Crystal structure of the double mutant Y88H-P104H of a SOD-like protein from Bacillus subtilis. | | Descriptor: | COPPER (II) ION, Hypothetical superoxide dismutase-like protein yojM, ZINC ION | | Authors: | Calderone, V, Mangani, S, Banci, L, Benvenuti, M, Bertini, I, Fantoni, A, Viezzoli, M.S. | | Deposit date: | 2004-10-22 | | Release date: | 2005-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | From an Inactive Prokaryotic SOD Homologue to an Active Protein through Site-Directed Mutagenesis.
J.Am.Chem.Soc., 127, 2005
|
|
3A85
 
 | |
3A80
 
 | |
6WJD
 
 | |
3A7X
 
 | |
2C8L
 
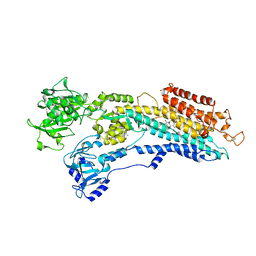 | | Crystal Structure of (SR) Calcium-ATPase E2(Tg) form | | Descriptor: | OCTANOIC ACID [3S-[3ALPHA, 3ABETA, 4ALPHA, ... | | Authors: | Jensen, A.M, Sorensen, T.L, Olesen, C, Moller, J.V, Nissen, P. | | Deposit date: | 2005-12-06 | | Release date: | 2006-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Modulatory and Catalytic Modes of ATP Binding by the Calcium Pump
Embo J., 25, 2006
|
|
5G5J
 
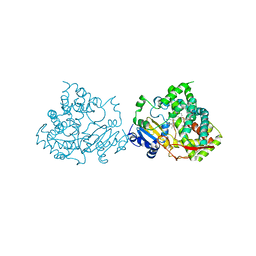 | | Crystal structure of human CYP3A4 bound to metformin | | Descriptor: | CYTOCHROME P450 3A4, Metformin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I. | | Deposit date: | 2016-05-25 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Heme Binding Biguanides Target Cytochrome P450-Dependent Cancer Cell Mitochondria.
Cell Chem Biol, 24, 2017
|
|
2M9A
 
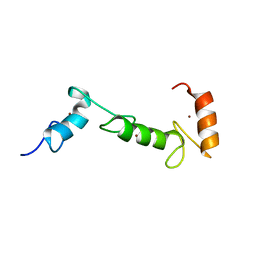 | | Solution NMR Structure of E3 ubiquitin-protein ligase ZFP91 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR7784A | | Descriptor: | E3 ubiquitin-protein ligase ZFP91, ZINC ION | | Authors: | Pederson, K, Shastry, R, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-05 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Hr7784A
To be Published
|
|
1XTL
 
 | | Crystal structure of P104H mutant of SOD-like protein from Bacillus subtilis. | | Descriptor: | COPPER (II) ION, Hypothetical superoxide dismutase-like protein yojM, ZINC ION | | Authors: | Calderone, V, Mangani, S, Banci, L, Benvenuti, M, Bertini, I, Viezzoli, M.S, Fantoni, A. | | Deposit date: | 2004-10-22 | | Release date: | 2005-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | From an Inactive Prokaryotic SOD Homologue to an Active Protein through Site-Directed Mutagenesis.
J.Am.Chem.Soc., 127, 2005
|
|
4BFD
 
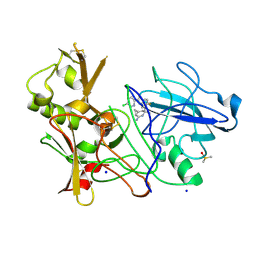 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH CHEMICAL LIGAND | | Descriptor: | BETA-SECRETASE 1, DIMETHYL SULFOXIDE, N-[3-[(1S,3S,6S)-5-azanyl-3-methyl-4-azabicyclo[4.1.0]hept-4-en-3-yl]-4-fluoranyl-phenyl]-5-chloranyl-pyridine-2-carbox amide, ... | | Authors: | Banner, D.W, Benz, J, Stihle, M. | | Deposit date: | 2013-03-18 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bace1 Inhibitors: A Head Group Scan on a Series of Amides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4BEK
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH CHEMICAL LIGAND | | Descriptor: | (4S)-4-(4-methoxyphenyl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine, BETA-SECRETASE 1, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D.W, Benz, J, Stihle, M. | | Deposit date: | 2013-03-11 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Bace1 Inhibitors: A Head Group Scan on a Series of Amides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3A81
 
 | |
3A0B
 
 | | Crystal structure of Br-substituted Photosystem II complex | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 5-[(2E,6E,10E,14E,18E,22E)-3,7,11,15,19,23,27-HEPTAMETHYLOCTACOSA-2,6,10,14,18,22,26-HEPTAENYL]-2,3-DIMETHYLBENZO-1,4-QUINONE, ... | | Authors: | Kawakami, K, Umena, Y, Kamiya, N, Shen, J.-R. | | Deposit date: | 2009-03-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Location of chloride and its possible functions in oxygen-evolving photosystem II revealed by X-ray crystallography
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3A0H
 
 | | Crystal structure of I-substituted Photosystem II complex | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 5-[(2E,6E,10E,14E,18E,22E)-3,7,11,15,19,23,27-HEPTAMETHYLOCTACOSA-2,6,10,14,18,22,26-HEPTAENYL]-2,3-DIMETHYLBENZO-1,4-QUINONE, ... | | Authors: | Kawakami, K, Umena, Y, Kamiya, N, Shen, J.-R. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Location of chloride and its possible functions in oxygen-evolving photosystem II revealed by X-ray crystallography
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2A0N
 
 | |
3A89
 
 | |
3A7Y
 
 | |
1KGI
 
 | | Rat transthyretin (also called prealbumin) complex with 3,3',5,5'-tetraiodothyroacetic acid (t4ac) | | Descriptor: | 3,3',5,5'-TETRAIODOTHYROACETIC ACID, TRANSTHYRETIN | | Authors: | Wojtczak, A, Neumann, P, Muziol, T, Cody, V, Luft, J.R, Pangborn, W. | | Deposit date: | 2001-11-27 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complex of rat transthyretin with tetraiodothyroacetic acid refined at 2.1 and 1.8 A resolution.
Acta Biochim.Pol., 48, 2001
|
|
3A7Z
 
 | |
3A8B
 
 | |
3A84
 
 | |
3AUS
 
 | |
2KXQ
 
 | | Solution Structure of Smurf2 WW2 and WW3 bound to Smad7 PY motif containing peptide | | Descriptor: | E3 ubiquitin-protein ligase SMURF2, Smad7 PY motif containing peptide | | Authors: | Chong, A, Lin, H, Wrana, J, Forman-Kay, J.D. | | Deposit date: | 2010-05-11 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Coupling of tandem Smad ubiquitination regulatory factor (Smurf) WW domains modulates target specificity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1BE7
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN C42S MUTANT | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Maher, M, Guss, J.M, Wilce, M, Wedd, A.G. | | Deposit date: | 1998-05-20 | | Release date: | 1998-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Rubredoxin from Clostridium Pasteurianum: Mutation of the Iron Cysteinyl Ligands to Serine. Crystal and Molecular Structures of the Oxidised and Dithionite-Treated Forms of the Cys42Ser Mutant
J.Am.Chem.Soc., 120, 1998
|
|
