1W1I
 
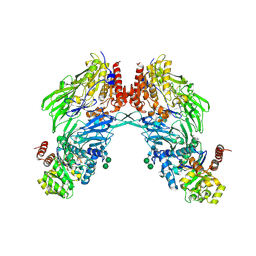 | | Crystal structure of dipeptidyl peptidase IV (DPPIV or CD26) in complex with adenosine deaminase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weihofen, W.A, Liu, J, Reutter, W, Saenger, W, Fan, H. | | Deposit date: | 2004-06-22 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Crystal structure of CD26/dipeptidyl-peptidase IV in complex with adenosine deaminase reveals a highly amphiphilic interface.
J. Biol. Chem., 279, 2004
|
|
7ZZ5
 
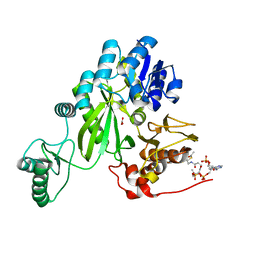 | | Cryo-EM structure of "BC open" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, BICARBONATE ION, Pyruvate carboxylase | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZYZ
 
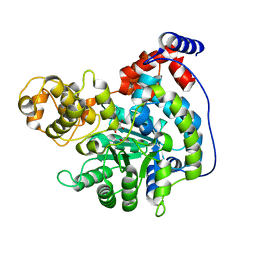 | | Cryo-EM structure of "CT oxa" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MANGANESE (II) ION, OXALOACETATE ION, Pyruvate carboxylase | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
4PEZ
 
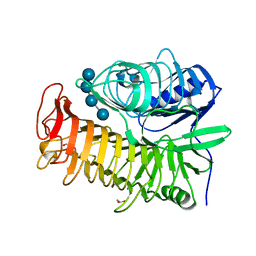 | | Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
7ZZ1
 
 | | Cryo-EM structure of "CT react" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | BIOTIN, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ2
 
 | | Cryo-EM structure of "CT pyr" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, PYRUVIC ACID, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ0
 
 | | Cryo-EM structure of "CT empty" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Pyruvate carboxylase | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
1LUL
 
 | | DB58, A LEGUME LECTIN FROM DOLICHOS BIFLORUS | | Descriptor: | CALCIUM ION, LECTIN DB58, MANGANESE (II) ION | | Authors: | Hamelryck, T.W, Bouckaert, J, Dao-Thi, M.H, Wyns, L, Etzler, M, Loris, R. | | Deposit date: | 1998-06-30 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Carbohydrate binding, quaternary structure and a novel hydrophobic binding site in two legume lectin oligomers from Dolichos biflorus.
J.Mol.Biol., 286, 1999
|
|
7ZZ4
 
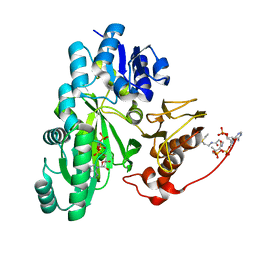 | | Cryo-EM structure of "BC closed" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
4UP1
 
 | |
8PT4
 
 | | beta-Ureidopropionase tetramer | | Descriptor: | Beta-ureidopropionase | | Authors: | Cederfelt, D, Dobritzsch, D. | | Deposit date: | 2023-07-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | The Allosteric Regulation of Beta-Ureidopropionase Depends on Fine-Tuned Stability of Active-Site Loops and Subunit Interfaces.
Biomolecules, 13, 2023
|
|
5XSJ
 
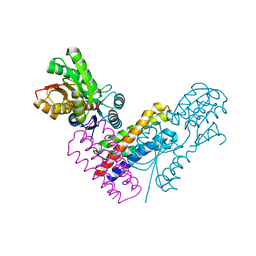 | | XylFII-LytSN complex | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, Signal transduction histidine kinase, LytS, ... | | Authors: | Li, J.X, Wang, C.Y, Zhang, P. | | Deposit date: | 2017-06-14 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7ZZ6
 
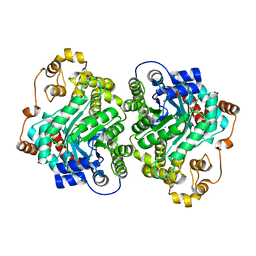 | | Cryo-EM structure of "CT-CT dimer" of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, PYRUVIC ACID, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
2VGS
 
 | | Crystal structure of E53QbsSHMT internal aldimine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PYRIDOXAL-5'-PHOSPHATE, SERINE HYDROXYMETHYLTRANSFERASE | | Authors: | Rajaram, V, Bhavani, B.S, Kaul, P, Prakash, V, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2007-11-15 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure Determination and Biochemical Studies on Bacillus Stearothermophilus E53Q Serine Hydroxymethyltransferase and its Complexes Provide Insights on Function and Enzyme Memory
FEBS J., 274, 2007
|
|
4P3G
 
 | |
7KC2
 
 | | Symmetry in Yeast Alcohol Dehydrogenase 1 -Closed Form with NADH | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Subramanian, R, Chang, L, Li, Z, Plapp, B.V. | | Deposit date: | 2020-10-04 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Cryo-Electron Microscopy Structures of Yeast Alcohol Dehydrogenase.
Biochemistry, 60, 2021
|
|
4P6Z
 
 | |
1MW5
 
 | | Structure of HI1480 from Haemophilus influenzae | | Descriptor: | HYPOTHETICAL PROTEIN HI1480 | | Authors: | Lim, K, Sarikaya, E, Howard, A, Galkin, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-09-27 | | Release date: | 2003-11-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel structure and nucleotide binding properties of HI1480 from Haemophilus influenzae: a protein with no known sequence homologues
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
6UOH
 
 | | Asparaginase II from Escherichia coli | | Descriptor: | ASPARTIC ACID, L-asparaginase 2 | | Authors: | Araujo, T.S, Almeida, M.S, Lima, L.M.T.R. | | Deposit date: | 2019-10-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biophysical characterization of two commercially available preparations of the drug containing Escherichia coli L-Asparaginase 2.
Biophys.Chem., 271, 2021
|
|
4I08
 
 | | Crystal structure of beta-ketoacyl-acyl carrier protein reductase (FabG) from Vibrio cholerae in complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] reductase FabG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Osinski, T, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-16 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
4PDJ
 
 | | Neutron crystal Structure of E.coli Dihydrofolate Reductase complexed with folate and NADP+ | | Descriptor: | DIHYDROFOLIC ACID, Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Wan, Q, Kovalevsky, A.Y, Wilson, M, Langan, P, Dealwis, C, Bennett, B. | | Deposit date: | 2014-04-18 | | Release date: | 2015-04-15 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.599 Å), X-RAY DIFFRACTION | | Cite: | Toward resolving the catalytic mechanism of dihydrofolate reductase using neutron and ultrahigh-resolution X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4L5I
 
 | |
3O4X
 
 | |
4PSM
 
 | | Crystal structure of pfuThermo-DBP-RP1 (crystal form II) | | Descriptor: | SULFATE ION, ssDNA binding protein | | Authors: | Gahlei, H, von Moeller, H, Eppers, D, Loll, B, Wahl, M.C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Entrapment of DNA in an intersubunit tunnel system of a single-stranded DNA-binding protein.
Nucleic Acids Res., 42, 2014
|
|
8J5Z
 
 | | The cryo-EM structure of the TwOSC1 tetramer | | Descriptor: | Terpene cyclase/mutase family member, octyl beta-D-glucopyranoside | | Authors: | Ma, X, Yuru, T, Yunfeng, L, Jiang, T. | | Deposit date: | 2023-04-24 | | Release date: | 2023-11-01 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | Structural and Catalytic Insight into the Unique Pentacyclic Triterpene Synthase TwOSC.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
