5N8M
 
 | |
8JPS
 
 | | Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map) | | Descriptor: | Atypical chemokine receptor 1, C-C motif chemokine 7 | | Authors: | Banerjee, R, Khanppnavar, B, Maharana, J, Saha, S, Korkhov, V.M, Shukla, A.K. | | Deposit date: | 2023-06-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Molecular mechanism of distinct chemokine engagement and functional divergence of the human Duffy antigen receptor.
Cell, 187, 2024
|
|
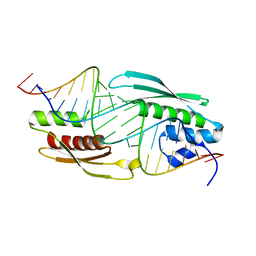
7JMM
 
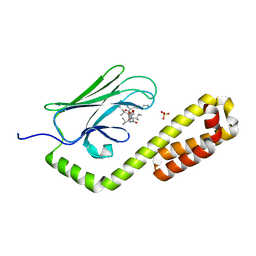 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RAKNIILLSR | | Descriptor: | Alkaline phosphatase, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2020-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JMU
 
 | | Hen egg-white lysozyme with ionic liquid ethylammonium nitrate | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Han, Q, Darmanin, C, Smith, K, Greaves, T, Drummond, C. | | Deposit date: | 2020-08-02 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Lysozyme conformational changes with ionic liquids: Spectroscopic, small angle x-ray scattering and crystallographic study.
J Colloid Interface Sci, 585, 2021
|
|
8K8M
 
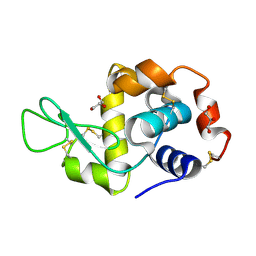 | |
7JN8
 
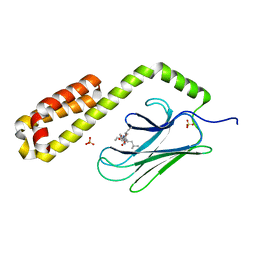 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RGNTLVIVSR | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5NBJ
 
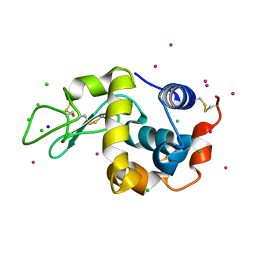 | | DLS Tetragonal - ReHEWL | | Descriptor: | CHLORIDE ION, Lysozyme C, RHENIUM, ... | | Authors: | Brink, A, Helliwell, J.R. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.266 Å) | | Cite: | New leads for fragment-based design of rhenium/technetium radiopharmaceutical agents.
IUCrJ, 4, 2017
|
|
5NCW
 
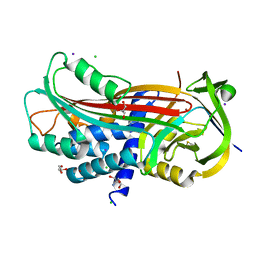 | | Structure of the trypsin induced serpin-type proteinase inhibitor, miropin (V367K/K368A mutant). | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
5NOJ
 
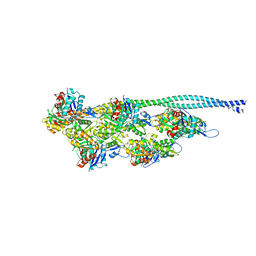 | | Ca2+-induced Movement of Tropomyosin on Native Cardiac Thin Filaments - "OPEN" state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Risi, C, Eisner, J, Belknap, B, Heeley, D.H, White, H.D, Schroeder, G.F, Galkin, V.E. | | Deposit date: | 2017-04-12 | | Release date: | 2017-08-02 | | Last modified: | 2017-08-09 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Ca(2+)-induced movement of tropomyosin on native cardiac thin filaments revealed by cryoelectron microscopy.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NP5
 
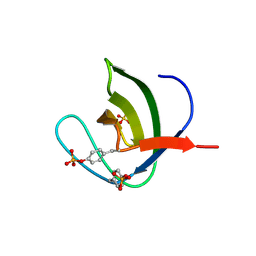 | | Abl2 SH3 pTyr116/161 | | Descriptor: | Abelson tyrosine-protein kinase 2, SULFATE ION | | Authors: | Mero, B, Radnai, L, Gogl, G, Leveles, I, Buday, L. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into the tyrosine phosphorylation-mediated inhibition of SH3 domain-ligand interactions.
J.Biol.Chem., 294, 2019
|
|
7JXZ
 
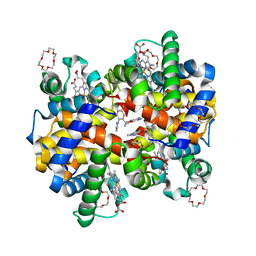 | | Structure of HbA with compound (S)-4 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3-{(1S)-1-[5-fluoro-2-(1H-pyrazol-1-yl)phenyl]ethoxy}-5-(3-methyl-1H-pyrazol-4-yl)pyridin-2-amine, CARBON MONOXIDE, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | PF-07059013: A Noncovalent Modulator of Hemoglobin for Treatment of Sickle Cell Disease.
J.Med.Chem., 64, 2021
|
|
5L3L
 
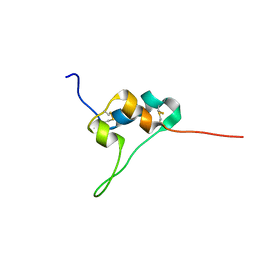 | | D11 bound IGF-II | | Descriptor: | Insulin-like growth factor II | | Authors: | Hexnerova, R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Probing Receptor Specificity by Sampling the Conformational Space of the Insulin-like Growth Factor II C-domain.
J.Biol.Chem., 291, 2016
|
|
5L3M
 
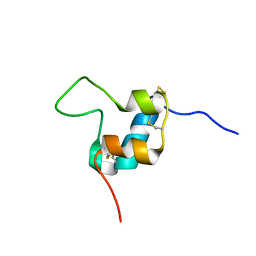 | | D11 bound [S39_PQ]-IGF-II | | Descriptor: | Insulin-like growth factor II | | Authors: | Hexnerova, R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Probing Receptor Specificity by Sampling the Conformational Space of the Insulin-like Growth Factor II C-domain.
J.Biol.Chem., 291, 2016
|
|
5L7M
 
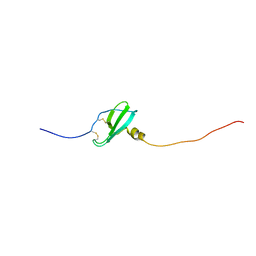 | | Murin CXCL13 solution structure | | Descriptor: | C-X-C motif chemokine 13 | | Authors: | Monneau, Y.R, Lortat-Jacob, H. | | Deposit date: | 2016-06-03 | | Release date: | 2017-06-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of CXCL13 and heparan sulfate binding show that GAG binding site and cellular signalling rely on distinct domains.
Open Biol, 7, 2017
|
|
5L3N
 
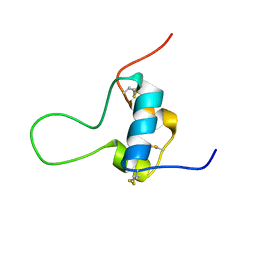 | | D11 bound [N29, S39_PQ]-IGF-II | | Descriptor: | Insulin-like growth factor II | | Authors: | Hexnerova, R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Probing Receptor Specificity by Sampling the Conformational Space of the Insulin-like Growth Factor II C-domain.
J.Biol.Chem., 291, 2016
|
|
8JJS
 
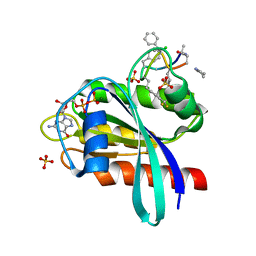 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor AP10343 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAA-ILE-SAR-SAR-7T2-SAR-IAE-LEU-MEA-MLE-7TK, ... | | Authors: | Irie, M, Fukami, T.A, Tanada, M, Ohta, A, Torizawa, T. | | Deposit date: | 2023-05-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Development of Orally Bioavailable Peptides Targeting an Intracellular Protein: From a Hit to a Clinical KRAS Inhibitor.
J.Am.Chem.Soc., 145, 2023
|
|
7JN1
 
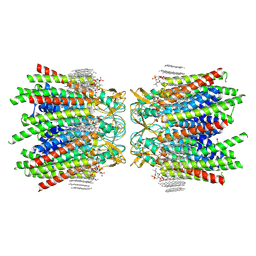 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-08-03 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
5LFT
 
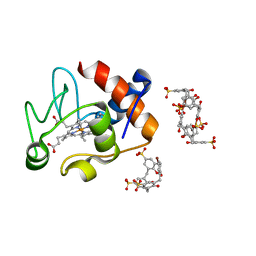 | | Crystal structure of cytochrome c - Bromo-trisulfonatocalix[4]arene complexes | | Descriptor: | BROMIDE ION, Bromo-trisulfonatocalix[4]arene, Cytochrome c iso-1, ... | | Authors: | Doolan, A.M, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2016-07-04 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | Protein Recognition by Functionalized Sulfonatocalix[4]arenes.
Chemistry, 24, 2018
|
|
8KFH
 
 | |
5LIE
 
 | |
5LJC
 
 | | Crystal structure of holo human CRBP1 | | Descriptor: | RETINOL, Retinol-binding protein 1, SODIUM ION | | Authors: | Zanotti, G, Vallese, F, Berni, R, Menozzi, I. | | Deposit date: | 2016-07-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.433 Å) | | Cite: | Structural and molecular determinants affecting the interaction of retinol with human CRBP1.
J. Struct. Biol., 197, 2017
|
|
5LJH
 
 | | Crystal structure of human apo CRBP1/K40L mutant | | Descriptor: | Retinol-binding protein 1, SODIUM ION | | Authors: | Zanotti, G, Vallese, F, Berni, R, Menozzi, I. | | Deposit date: | 2016-07-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and molecular determinants affecting the interaction of retinol with human CRBP1.
J. Struct. Biol., 197, 2017
|
|
5LH4
 
 | | Trypsin inhibitors for the treatment of pancreatitis - cpd 1 | | Descriptor: | (2~{S},4~{S})-1-[4-(aminomethyl)phenyl]carbonyl-4-(4-cyclopropyl-1,2,3-triazol-1-yl)-~{N}-(2,2-diphenylethyl)pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiering, N, D'Arcy, A, Skaanderup, P, Simic, O, Brandl, T, Woelcke, J. | | Deposit date: | 2016-07-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Trypsin inhibitors for the treatment of pancreatitis.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
8KDU
 
 | | Crystal structure of trypsin with its substrate | | Descriptor: | (2~{R})-2-benzamido-5-carbamimidamido-pentanoic acid, ARGININE, CALCIUM ION, ... | | Authors: | Akbar, Z, Ahmad, M.S. | | Deposit date: | 2023-08-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Michaelis complex of trypsin with N-alpha-benzoyl-l-arginine ethyl ester
J Chin Chem Soc, n/a, 2024
|
|
8KFJ
 
 | |
