3ZTN
 
 | | STRUCTURE OF INFLUENZA A NEUTRALIZING ANTIBODY SELECTED FROM CULTURES OF SINGLE HUMAN PLASMA CELLS IN COMPLEX WITH HUMAN H1 INFLUENZA HAEMAGGLUTININ. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FI6V3 ANTIBODY LIGHT CHAIN, ... | | Authors: | Hubbard, P.A, Ritchie, A.J, Corti, D, Voss, J.E, Gamblin, S.J, Codoni, G, Macagno, A, Jarrossay, D, Pinna, D, Minola, A, Vanzetta, F, Silacci, C, Fernandez-Rodriguez, B.M, Agatic, G, Giacchetto-Sasselli, I, Vachieri, S.G, Sallusto, F, Collins, P.J, Haire, L.F, Temperton, N, Langedijk, J.P.M, Skehel, J.J, Lanzavecchia, A. | | Deposit date: | 2011-07-12 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | A Neutralizing Antibody Selected from Plasma Cells that Binds to Group 1 and Group 2 Influenza a Hemagglutinins.
Science, 333, 2011
|
|
2QZ3
 
 | | Crystal structure of a glycoside hydrolase family 11 xylanase from Bacillus subtilis in complex with xylotetraose | | Descriptor: | ACETIC ACID, Endo-1,4-beta-xylanase A, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Vandermarliere, E, Bourgois, T.M, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | Deposit date: | 2007-08-16 | | Release date: | 2007-12-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
1Z4N
 
 | | Structure of beta-phosphoglucomutase with inhibitor bound alpha-galactose 1-phosphate cocrystallized with Fluoride | | Descriptor: | 1-O-phosphono-alpha-D-galactopyranose, Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Tremblay, L.W, Zhang, G, Dai, J, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2005-03-16 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Chemical Confirmation of a Pentavalent Phosphorane in Complex with beta-Phosphoglucomutase
J.Am.Chem.Soc., 127, 2005
|
|
4DKE
 
 | | Crystal Structure of Human Interleukin-34 Bound to FAb1.1 | | Descriptor: | FAb1.1 Heavy Chain, FAb1.1 Light Chain, Interleukin-34, ... | | Authors: | Ma, X, Chen, Y, Stawicki, S, Wu, Y, Bazan, J.F, Starovasnik, M.A. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for the Dual Recognition of Helical Cytokines IL-34 and CSF-1 by CSF-1R.
Structure, 20, 2012
|
|
2AOI
 
 | | Crystal structure analysis of HIV-1 protease with a substrate analog P1-P6 | | Descriptor: | PEPTIDE INHIBITOR, POL POLYPROTEIN, SULFATE ION | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2ZKN
 
 | |
1S6Q
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R147681 | | Descriptor: | 4-[4-(2,4,6-TRIMETHYL-PHENYLAMINO)-PYRIMIDIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
3BQD
 
 | | Doubling the Size of the Glucocorticoid Receptor Ligand Binding Pocket by Deacylcortivazol | | Descriptor: | 1-[(1R,2R,3aS,3bS,10aR,10bS,11S,12aS)-1,11-dihydroxy-2,5,10a,12a-tetramethyl-7-phenyl-1,2,3,3a,3b,7,10,10a,10b,11,12,12a-dodecahydrocyclopenta[5,6]naphtho[1,2-f]indazol-1-yl]-2-hydroxyethanone, Glucocorticoid receptor, Nuclear receptor coactivator 1 | | Authors: | Xu, H.E. | | Deposit date: | 2007-12-20 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Doubling the size of the glucocorticoid receptor ligand binding pocket by deacylcortivazol.
Mol.Cell.Biol., 28, 2008
|
|
2QSC
 
 | | Crystal structure analysis of anti-HIV-1 V3-Fab F425-B4e8 in complex with a V3-peptide | | Descriptor: | CHLORIDE ION, Envelope glycoprotein gp120, Fab F425-B4e8, ... | | Authors: | Bell, C.H, Schiefner, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2007-07-30 | | Release date: | 2008-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of antibody F425-B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization.
J.Mol.Biol., 375, 2008
|
|
3OD8
 
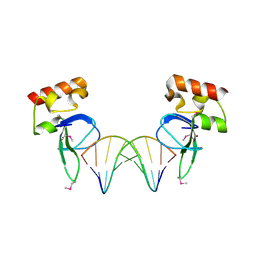 | | Human PARP-1 zinc finger 1 (Zn1) bound to DNA | | Descriptor: | 5'-D(*CP*CP*CP*AP*AP*GP*CP*GP*GP*C)-3', 5'-D(*GP*CP*CP*GP*CP*TP*TP*GP*GP*G)-3', Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Langelier, M.-F. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Poly(ADP-ribose) Polymerase-1 (PARP-1) Zinc Fingers Bound to DNA: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO DNA-DEPENDENT PARP-1 ACTIVITY.
J.Biol.Chem., 286, 2011
|
|
2JMW
 
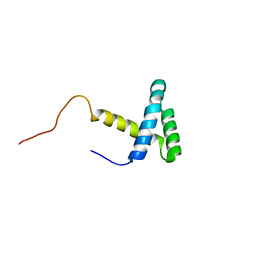 | | Structure of DNA-Binding Domain of Arabidopsis GT-1 | | Descriptor: | DNA binding protein GT-1 | | Authors: | Nagata, T, Niyada, E, Noto, K, Ikeda, Y, Yamamoto, Y, Uesugi, S, Murata, J, Hiratsuka, K, Katahira, M. | | Deposit date: | 2006-12-11 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: mechanism for an increase in DNA-binding affinity through phosphorylation.
Proteins, 78, 2010
|
|
3SX9
 
 | |
3ODE
 
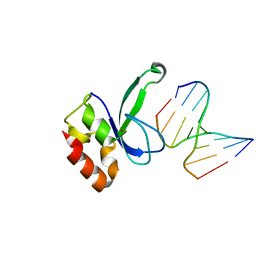 | | Human PARP-1 zinc finger 2 (Zn2) bound to DNA | | Descriptor: | 5'-D(*CP*CP*CP*AP*AP*GP*CP*G)-3', 5'-D(*CP*GP*CP*TP*TP*GP*GP*G)-3', Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Langelier, M.-F. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structures of Poly(ADP-ribose) Polymerase-1 (PARP-1) Zinc Fingers Bound to DNA: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO DNA-DEPENDENT PARP-1 ACTIVITY.
J.Biol.Chem., 286, 2011
|
|
3ODC
 
 | | Human PARP-1 zinc finger 2 (Zn2) bound to DNA | | Descriptor: | 5'-D(*CP*CP*CP*AP*GP*AP*CP*G)-3', 5'-D(*CP*GP*TP*CP*TP*GP*GP*G)-3', Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Langelier, M.-F. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Poly(ADP-ribose) Polymerase-1 (PARP-1) Zinc Fingers Bound to DNA: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO DNA-DEPENDENT PARP-1 ACTIVITY.
J.Biol.Chem., 286, 2011
|
|
1K7C
 
 | |
2FXU
 
 | | X-ray Structure of Bistramide A- Actin Complex at 1.35 A resolution. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rizvi, S.A, Tereshko, V, Kossiakoff, A.A, Kozmin, S.A. | | Deposit date: | 2006-02-06 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of bistramide a-actin complex at a 1.35 A resolution
J.Am.Chem.Soc., 128, 2006
|
|
2AOH
 
 | | Crystal structure analysis of HIV-1 Protease mutant V82A with a substrate analog P6-PR | | Descriptor: | CHLORIDE ION, PEPTIDE INHIBITOR, POL POLYPROTEIN, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
3OPM
 
 | | Crystal Structure of Human DPP4 Bound to TAK-294 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[3-(aminomethyl)-2-(2-methylpropyl)-1-oxo-4-phenyl-1,2-dihydroisoquinolin-6-yl]oxy}acetamide, ... | | Authors: | Yano, J.K, Aertgeerts, K. | | Deposit date: | 2010-09-01 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Identification of 3-aminomethyl-1,2-dihydro-4-phenyl-1-isoquinolones: a new class of potent, selective, and orally active non-peptide dipeptidyl peptidase IV inhibitors that form a unique interaction with Lys554.
Bioorg.Med.Chem., 19, 2011
|
|
2BAN
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with JANSSEN-R157208 | | Descriptor: | 5-ETHYL-3-[(2-METHOXYETHYL)METHYLAMINO]-6-METHYL-4-(3-METHYLBENZYL)PYRIDIN-2(1H)-ONE, MANGANESE (II) ION, Reverse transcriptase P51 subunit, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structures for HIV-1 Reverse Transcriptase in Complexes with Three Pyridinone Derivatives: A New Class of Non-Nucleoside Inhibitors Effective against a Broad Range of Drug-Resistant Strains.
J.Med.Chem., 48, 2005
|
|
2L6E
 
 | | NMR Structure of the monomeric mutant C-terminal domain of HIV-1 Capsid in complex with stapled peptide Inhibitor | | Descriptor: | Capsid protein p24, NYAD-13 stapled peptide inhibitor | | Authors: | Bhattacharya, S, Zhang, H, Debnath, A.K, Cowburn, D. | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a hydrocarbon stapled peptide inhibitor in complex with monomeric C-terminal domain of HIV-1 capsid.
J.Biol.Chem., 283, 2008
|
|
2LCU
 
 | | NMR structure of BC28.1 | | Descriptor: | Bc28.1 | | Authors: | Roumestand, C, Delbecq, S, Yang, Y. | | Deposit date: | 2011-05-09 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of Bc28.1, Major Erythrocyte-binding Protein from Babesia canis Merozoite Surface.
J.Biol.Chem., 287, 2012
|
|
2LRS
 
 | | The second dsRBD domain from A. thaliana DICER-LIKE 1 | | Descriptor: | Endoribonuclease Dicer homolog 1 | | Authors: | Burdisso, P, Suarez, I, Bersch, B, Bologna, N, Palatnik, J, Boisbouvier, J, Rasia, R. | | Deposit date: | 2012-04-12 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Second Double-Stranded RNA Binding Domain of Dicer-like Ribonuclease 1: Structural and Biochemical Characterization.
Biochemistry, 51, 2012
|
|
2IRT
 
 | |
1HQL
 
 | | The xenograft antigen in complex with the B4 isolectin of Griffonia simplicifolia lectin-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LECTIN, ... | | Authors: | Tempel, W, Lipscomb, L.A, Rose, J.P, Woods, R.J. | | Deposit date: | 2000-12-18 | | Release date: | 2002-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The xenograft antigen bound to Griffonia simplicifolia lectin 1-B(4). X-ray crystal structure of the complex and molecular dynamics characterization of the binding site.
J.Biol.Chem., 277, 2002
|
|
1D4K
 
 | | HIV-1 PROTEASE COMPLEXED WITH A MACROCYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | HIV-1 PROTEASE, N-13-[(10S,13S)-9,12-DIOXO-10-(2-BUTYL)-2-OXA-8,11-DIAZABICYCLO [13.2.2] NONADECA-15,17,18-TRIENE] (2R)-BENZYL-(4S)-HYDROXY-5-AMINOPENTANOIC (1R)-HYDROXY-(2S)-INDANEAMIDE, SULFATE ION | | Authors: | Tyndall, J.D, Reid, R.C, Tyssen, D.P, Jardine, D.K, Todd, B, Passmore, M, March, D.R, Pattenden, L.K, Alewood, D, Hu, S.H, Alewood, P.F, Birch, C.J, Martin, J.L, Fairlie, D.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease.
J.Med.Chem., 43, 2000
|
|
