1QC6
 
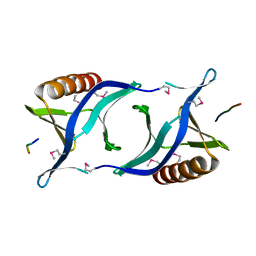 | | EVH1 domain from ENA/VASP-like protein in complex with ACTA peptide | | Descriptor: | EVH1 DOMAIN FROM ENA/VASP-LIKE PROTEIN, PHE-GLU-PHE-PRO-PRO-PRO-PRO-THR-ASP-GLU-GLU | | Authors: | Fedorov, A.A, Fedorov, E.V, Gertler, F.B, Almo, S.C. | | Deposit date: | 1999-05-17 | | Release date: | 1999-05-25 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of EVH1, a novel proline-rich ligand-binding module involved in cytoskeletal dynamics and neural function
Nat.Struct.Biol., 6, 1999
|
|
3RTW
 
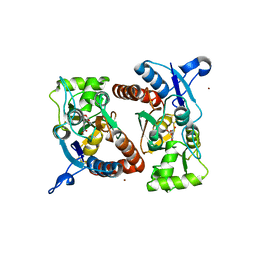 | |
2ZAL
 
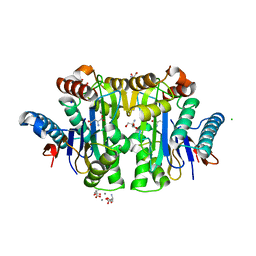 | | Crystal structure of E. coli isoaspartyl aminopeptidase/L-asparaginase in complex with L-aspartate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ASPARTIC ACID, CALCIUM ION, ... | | Authors: | Michalska, K, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2007-10-07 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of isoaspartyl aminopeptidase in complex with L-aspartate
J.Biol.Chem., 280, 2005
|
|
2VGK
 
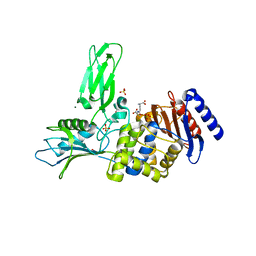 | | Crystal structure of Actinomadura R39 DD-peptidase complexed with a peptidoglycan-mimetic cephalosporin | | Descriptor: | (2R)-2-AMINO-7-{[(1R)-1-CARBOXYETHYL]AMINO}-7-OXOHEPTANOIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, MAGNESIUM ION, ... | | Authors: | Sauvage, E, kerff, F, Herman, R, Charlier, P. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures of Complexes of Bacterial Dd-Peptidases with Peptidoglycan-Mimetic Ligands: The Substrate Specificity Puzzle.
J.Mol.Biol., 381, 2008
|
|
7LZ2
 
 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with methionine | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LZ0
 
 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LZ1
 
 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with serine | | Descriptor: | GLYCEROL, Glutamate receptor 3.4, SERINE, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
2OKT
 
 | | Crystal structure of O-succinylbenzoic acid synthetase from Staphylococcus aureus, ligand-free form | | Descriptor: | O-succinylbenzoic acid synthetase | | Authors: | Patskovsky, Y, Toro, R, Malashkevich, V, Sauder, J.M, Ozyurt, S, Smith, D, Dickey, M, Maletic, M, Powell, A, Gheyi, T, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-17 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2D9Q
 
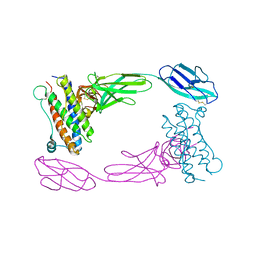 | | Crystal Structure of the Human GCSF-Receptor Signaling Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CSF3, Granulocyte colony-stimulating factor receptor | | Authors: | Tamada, T, Kuroki, R. | | Deposit date: | 2005-12-12 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6I9X
 
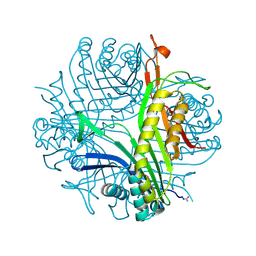 | | urate oxidase under 35 bar of argon | | Descriptor: | 8-AZAXANTHINE, ARGON, SODIUM ION, ... | | Authors: | Prange, T, Colloc'h, N. | | Deposit date: | 2018-11-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
7EIH
 
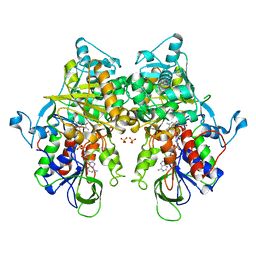 | | Ancestral L-Lys oxidase (ligand free form) | | Descriptor: | FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Sugiura, S, Nakano, S, Niwa, M, Hasebe, F, Ito, S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic mechanism of ancestral L-lysine oxidase assigned by sequence data mining.
J.Biol.Chem., 297, 2021
|
|
1Y1M
 
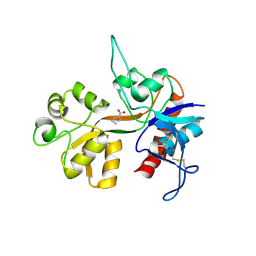 | |
1Y1Z
 
 | |
1ZZN
 
 | |
4Q9R
 
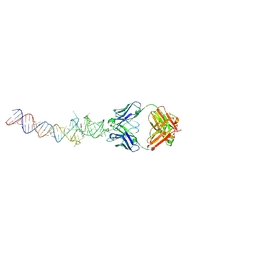 | | Crystal structure of an RNA aptamer bound to trifluoroethyl-ligand analog in complex with Fab | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Fab BL3-6, HEAVY CHAIN, ... | | Authors: | Huang, H, Suslov, N.B, Li, N.-S, Shelke, S.A, Evans, M.E, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-18 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
1DRM
 
 | | CRYSTAL STRUCTURE OF THE LIGAND FREE BJFIXL HEME DOMAIN | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SENSOR PROTEIN FIXL | | Authors: | Gong, W, Hao, B, Mansy, S.S, Gonzalez, G, Gilles-Gonzalez, M.A, Chan, M.K. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4A2Z
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 4-METHOXY-2,3,6-TRIMETHYL-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)BENZENESULFONAMIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Robinson, D.A, Brand, S, Fairlamb, A.H, Ferguson, M.A.J, Frearson, J.A, Wyatt, P.G. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
1FRT
 
 | |
1FP7
 
 | | MONOVALENT CATION BINDING SITES IN N10-FORMYLTETRAHYDROFOLATE SYNTHETASE FROM MOORELLA THERMOACETICA | | Descriptor: | FORMATE--TETRAHYDROFOLATE LIGASE, POTASSIUM ION, SULFATE ION | | Authors: | Radfar, R, Leaphart, A, Brewer, J.M, Minor, W, Odom, J.D. | | Deposit date: | 2000-08-30 | | Release date: | 2001-08-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Cation binding and thermostability of FTHFS monovalent cation binding sites and thermostability of N10-formyltetrahydrofolate synthetase from Moorella thermoacetica.
Biochemistry, 39, 2000
|
|
2P3W
 
 | |
6EFC
 
 | | Hsa Siglec + Unique domains (unliganded) | | Descriptor: | CALCIUM ION, SODIUM ION, Streptococcal hemagglutinin | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
7MHC
 
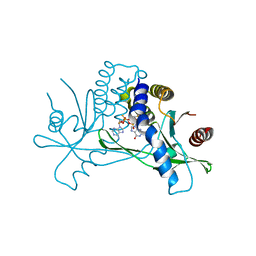 | | Structure of human STING in complex with MK-1454 | | Descriptor: | (2R,5R,7R,8S,10R,12aR,14R,15S,15aR,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-15,16-difluoro-2,10-bis(sulfanyl)octahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A kinase-cGAS cascade to synthesize a therapeutic STING activator.
Nature, 603, 2022
|
|
1GBV
 
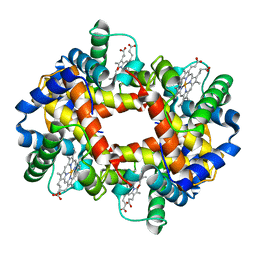 | | (ALPHA-OXY, BETA-(C112G)DEOXY) T-STATE HUMAN HEMOGLOBIN | | Descriptor: | HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vasquez, G.B, Ji, X, Pechik, I, Fronticelli, C, Gilliland, G.L. | | Deposit date: | 1995-12-20 | | Release date: | 1997-01-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cysteines beta93 and beta112 as probes of conformational and functional events at the human hemoglobin subunit interfaces.
Biophys.J., 76, 1999
|
|
1N4K
 
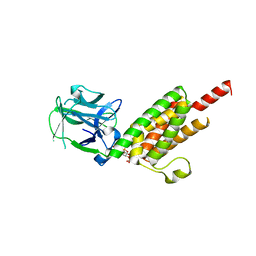 | | Crystal structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with IP3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Bosanac, I, Alattia, J.R, Mal, T.K, Chan, J, Talarico, S, Tong, F.K, Tong, K.I, Yoshikawa, F, Furuichi, T, Iwai, M, Michikawa, T, Mikoshiba, K, Ikura, M. | | Deposit date: | 2002-10-31 | | Release date: | 2002-12-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the inositol 1,4,5-trisphosphate receptor
binding core in complex with its ligand.
Nature, 420, 2002
|
|
4CP4
 
 | |
