1YUC
 
 | | Human Nuclear Receptor Liver Receptor Homologue-1, LRH-1, Bound to Phospholipid and a Fragment of Human SHP | | Descriptor: | GLYCEROL, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor 0B2, ... | | Authors: | Ortlund, E.A, Yoonkwang, L, Solomon, I.H, Hager, J.M, Safi, R, Choi, Y, Guan, Z, Tripathy, A, Raetz, C.R.H, McDonnell, D.P, Moore, D.D, Redinbo, M.R. | | Deposit date: | 2005-02-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP
Nat.Struct.Mol.Biol., 12, 2005
|
|
6TWL
 
 | |
4K95
 
 | | Crystal Structure of Parkin | | Descriptor: | E3 ubiquitin-protein ligase parkin, ZINC ION | | Authors: | Seirafi, M, Menade, M, Sauve, V, Kozlov, G, Trempe, J.-F, Nagar, B, Gehring, K. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.499 Å) | | Cite: | Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Science, 340, 2013
|
|
3S3V
 
 | | human dihydrofolate reductase Q35K/N64F double mutant binary complex with trimethoprim | | Descriptor: | Dihydrofolate reductase, SULFATE ION, TRIMETHOPRIM | | Authors: | Cody, V, Pace, J, Piraino, J, Queener, S.F. | | Deposit date: | 2011-05-18 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystallographic analysis reveals a novel second binding site for trimethoprim in active site double mutants of human dihydrofolate reductase.
J.Struct.Biol., 176, 2011
|
|
4HH3
 
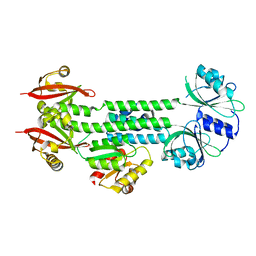 | | Structure of the AppA-PpsR2 core complex from Rb. sphaeroides | | Descriptor: | AppA protein, Transcriptional regulator, PpsR | | Authors: | Winkler, A, Heintz, U, Lindner, R, Reinstein, J, Shoeman, R, Schlichting, I. | | Deposit date: | 2012-10-09 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A ternary AppA-PpsR-DNA complex mediates light regulation of photosynthesis-related gene expression.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3DCT
 
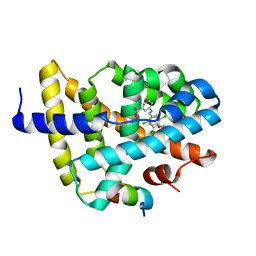 | | FXR with SRC1 and GW4064 | | Descriptor: | 3-[(E)-2-(2-chloro-4-{[3-(2,6-dichlorophenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}phenyl)ethenyl]benzoic acid, Bile acid receptor, Nuclear receptor coactivator 1 | | Authors: | Williams, S.P, Madauss, K.P. | | Deposit date: | 2008-06-04 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformationally constrained farnesoid X receptor (FXR) agonists: Naphthoic acid-based analogs of GW 4064.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3DCU
 
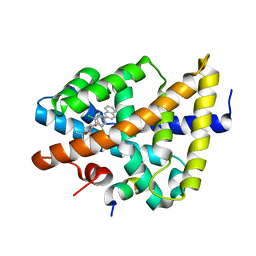 | | FXR with SRC1 and GSK8062 | | Descriptor: | 6-(4-{[3-(2,6-dichlorophenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}phenyl)naphthalene-1-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1 | | Authors: | Williams, S.P, Madauss, K.P. | | Deposit date: | 2008-06-04 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Conformationally constrained farnesoid X receptor (FXR) agonists: Naphthoic acid-based analogs of GW 4064.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4MEA
 
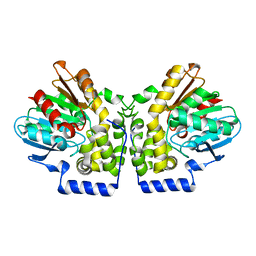 | |
4MEB
 
 | | Crystal structure of aCif-D158S | | Descriptor: | GLYCEROL, PHOSPHATE ION, Predicted protein | | Authors: | Bridges, A.A, Bahl, C.D, Madden, D.R. | | Deposit date: | 2013-08-25 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Signature motifs identify an acinetobacter cif virulence factor with epoxide hydrolase activity.
J.Biol.Chem., 289, 2014
|
|
2BJ3
 
 | | NIKR-apo | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NICKEL RESPONSIVE REGULATOR | | Authors: | Tahirov, T.H. | | Deposit date: | 2005-01-28 | | Release date: | 2005-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Pyrococcus Horikoshii Nikr: Nickel Sensing and Implications for the Regulation of DNA Recognition
J.Mol.Biol., 348, 2005
|
|
6BW4
 
 | | Crystal structure of RBBP4 in complex with PRDM16 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, PR domain zinc finger protein 16, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
2CKL
 
 | | Ring1b-Bmi1 E3 catalytic domain structure | | Descriptor: | IODIDE ION, POLYCOMB GROUP RING FINGER PROTEIN 4, UBIQUITIN LIGASE PROTEIN RING2, ... | | Authors: | Buchwald, G, van der Stoop, P, Weichenrieder, O, Perrakis, A, van Lohuizen, M, Sixma, T.K. | | Deposit date: | 2006-04-20 | | Release date: | 2006-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and E3-Ligase Activity of the Ring-Ring Complex of Polycomb Proteins Bmi1 and Ring1B.
Embo J., 25, 2006
|
|
3NAR
 
 | | Crystal structure of ZHX1 HD4 (zinc-fingers and homeoboxes protein 1, homeodomain 4) | | Descriptor: | SULFATE ION, Zinc fingers and homeoboxes protein 1 | | Authors: | Ren, J, Bird, L.E, Owens, R.J, Stammers, D.K, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel structural features in two ZHX homeodomains derived from a systematic study of single and multiple domains
Bmc Struct.Biol., 10, 2010
|
|
2BJ7
 
 | |
2BJ1
 
 | | NIKR IN OPEN CONFORMATION AND NICKEL BOUND TO HIGH-AFFINITY SITES | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, NICKEL RESPONSIVE REGULATOR | | Authors: | Tahirov, T.H. | | Deposit date: | 2005-01-27 | | Release date: | 2005-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Pyrococcus Horikoshii Nikr: Nickel Sensing and Implications for the Regulation of DNA Recognition
J.Mol.Biol., 348, 2005
|
|
2CAJ
 
 | | NikR from Helicobacter pylori in closed trans-conformation and nickel bound to 4 intermediary sites | | Descriptor: | CHLORIDE ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Dian, C, Schauer, K, Kapp, U, McSweeney, S.M, Labigne, A, Terradot, L. | | Deposit date: | 2005-12-21 | | Release date: | 2006-07-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of the Nickel Response in Helicobacter Pylori: Crystal Structures of Hpnikr in Apo and Nickel-Bound States.
J.Mol.Biol., 361, 2006
|
|
2B1D
 
 | |
3EZT
 
 | | Crystal Structure Analysis of Human HDAC8 D101E Variant | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, BETA-MERCAPTOETHANOL, Histone deacetylase 8, ... | | Authors: | Dowling, D.P, Gantt, S.L, Gattis, S.G, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2008-10-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors.
Biochemistry, 47, 2008
|
|
6Y17
 
 | |
1RV4
 
 | | E75L MUTANT OF RABBIT CYTOSOLIC SERINE HYDROXYMETHYLTRANSFERASE | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, ... | | Authors: | Szebenyi, D.M, Musayev, F.N, Di Salvo, M.L, Safo, M.K, Schirch, V. | | Deposit date: | 2003-12-12 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Serine Hydroxymethyltransferase: Role of Glu75 and Evidence that Serine Is Cleaved by a Retroaldol Mechanism.
Biochemistry, 43, 2004
|
|
1RVY
 
 | | E75Q MUTANT OF RABBIT CYTOSOLIC SERINE HYDROXYMETHYLTRANSFERASE, COMPLEX WITH GLYCINE | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Szebenyi, D.M, Musayev, F.N, Di Salvo, M.L, Safo, M.K, Schirch, V. | | Deposit date: | 2003-12-15 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Serine Hydroxymethyltransferase: Role of Glu75 and Evidence that Serine Is Cleaved by a Retroaldol Mechanism.
Biochemistry, 43, 2004
|
|
7BCB
 
 | |
3EZP
 
 | | Crystal Structure Analysis of human HDAC8 D101N variant | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, BETA-MERCAPTOETHANOL, Histone deacetylase 8, ... | | Authors: | Dowling, D.P, Gantt, S.L, Gattis, S.G, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2008-10-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors.
Biochemistry, 47, 2008
|
|
3F0R
 
 | | Crystal Structure Analysis of Human HDAC8 complexed with trichostatin A in a new monoclinic crystal form | | Descriptor: | Histone deacetylase 8, POTASSIUM ION, TRICHOSTATIN A, ... | | Authors: | Dowling, D.P, Gantt, S.L, Gattis, S.G, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2008-10-25 | | Release date: | 2008-12-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors.
Biochemistry, 47, 2008
|
|
3EW8
 
 | | Crystal Structure Analysis of human HDAC8 D101L variant | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Dowling, D.P, Gantt, S.L, Gattis, S.G, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2008-10-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors.
Biochemistry, 47, 2008
|
|
