5D7B
 
 | | Trigonal Crystal Structure of an acetylester hydrolase from Corynebacterium glutamicum | | Descriptor: | GLYCEROL, Homoserine O-acetyltransferase | | Authors: | Niefind, K, Toelzer, C, Pal, S, Altenbuchner, J, Watzlawick, H. | | Deposit date: | 2015-08-13 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A novel esterase subfamily with alpha / beta-hydrolase fold suggested by structures of two bacterial enzymes homologous to l-homoserine O-acetyl transferases.
Febs Lett., 590, 2016
|
|
5UGT
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT504 | | Descriptor: | 2-(2-chloranylphenoxy)-5-[(4-cyclopropyl-1,2,3-triazol-1-yl)methyl]phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
7Q20
 
 | | Ruminococcus gnavus ATC29149 endo-beta-1,4-galactosidase (RgGH98) in complex with blood group A trisaccharide | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Ruminococcus gnavus endogalactosidase GH98, ... | | Authors: | Owen, C.D, Wu, H, Crost, E.H, van Bakel, W, Gascuena, A.M, Latousakis, D, Hicks, T, Walpole, S, Urbanowicz, P.A, Ndeh, D, Monaco, S, Salom, L.S, Griffiths, R, Colvile, A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2021-10-22 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The human gut symbiont Ruminococcus gnavus shows specificity to blood group A antigen during mucin glycan foraging: Implication for niche colonisation in the gastrointestinal tract.
Plos Biol., 19, 2021
|
|
7N83
 
 | | Crystal Structure of SARS-CoV-2 NendoU in complex with Z2443429438 | | Descriptor: | (3S)-1-(phenylsulfonyl)pyrrolidin-3-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-12 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
8OI4
 
 | | Metagenomic Beta-galactosidase from Glycoside Hydrolase family GH154 | | Descriptor: | Beta-galactosidase, CHLORIDE ION, GLYCEROL | | Authors: | Pijning, T, Hameleers, L, Jurak, E, Guskov, A. | | Deposit date: | 2023-03-22 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Novel beta-galactosidase activity and first crystal structure of Glycoside Hydrolase family 154.
N Biotechnol, 80, 2023
|
|
7Z3X
 
 | | Crystal structure of FIR RRM1-2 Y115F mutant bound to FUSE ssDNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*T)-3'), DNA (5'-D(P*GP*TP*())-3'), ... | | Authors: | Ni, X, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-02 | | Release date: | 2022-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of FIR RRM1-2 Y115F mutant bound to FUSE ssDNA
To Be Published
|
|
6N6P
 
 | | Crystal structure of [FeFe]-hydrogenase in the presence of 7 mM Sodium dithionite | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Peters, J.W. | | Deposit date: | 2018-11-26 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
5UA4
 
 | |
5HGE
 
 | | Filamentous Assembly of Green Fluorescent Protein Supported by a C-terminal fusion of 18-residues, viewed in space group P212121 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Green fluorescent protein | | Authors: | Sawaya, M.R, Heller, D.M, McPartland, L, Hochschild, A, Eisenberg, D.S. | | Deposit date: | 2016-01-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Green Fluorescent Protein Fusion that Self Assembles as Polar Filaments
to be published
|
|
8RFZ
 
 | |
8OSB
 
 | | TWIST1-TCF4-ALX4 complex on specific DNA | | Descriptor: | DNA (25-MER), Homeobox protein aristaless-like 4, Transcription factor 4, ... | | Authors: | Morgunova, E, Kim, S, Popov, A, Wysocka, J, Taipale, J. | | Deposit date: | 2023-04-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | DNA-guided transcription factor cooperativity shapes face and limb mesenchyme.
Cell, 187, 2024
|
|
7VQB
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl pyrophosphate and dimethylallyl diphosphate | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, Di-trans-poly-cis-decaprenylcistransferase, FARNESYL DIPHOSPHATE, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
5HGM
 
 | | Hexameric HIV-1 CA in complex with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Capsid protein P24 | | Authors: | Jacques, D.A, James, L.C. | | Deposit date: | 2016-01-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis.
Nature, 536, 2016
|
|
7ZBV
 
 | |
7VQA
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with dimethylallyl diphosphate. | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, Di-trans-poly-cis-decaprenylcistransferase, MAGNESIUM ION, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
5UHL
 
 | | Crystal structure of the core catalytic domain of human O-GlcNAcase complexed with Thiamet G | | Descriptor: | (2Z,3aR,5R,6S,7R,7aR)-2-(ethylimino)-5-(hydroxymethyl)hexahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, O-GlcNAcase TIM-barrel domain, O-GlcNAcase stalk domain | | Authors: | Klein, D.J, Elsen, N.L. | | Deposit date: | 2017-01-11 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of the core catalytic domain of human O-GlcNAcase and molecular basis of activity and inhibition
To Be Published
|
|
7Z2J
 
 | | White Bream virus N7-Methyltransferase | | Descriptor: | Non-structural protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Shannon, A, Gauffre, P, Canard, B, Ferron, F. | | Deposit date: | 2022-02-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | A second type of N7-guanine RNA cap methyltransferase in an unusual locus of a large RNA virus genome.
Nucleic Acids Res., 50, 2022
|
|
7PDR
 
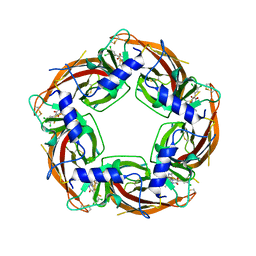 | | Crystal structure of Lymnaea stagnalis Acetylcholine-binding protein (Ls-AChBP) Q55R/M114V double mutant complexed with Dichloromezotiaz | | Descriptor: | 3-[3,5-bis(chloranyl)phenyl]-1-[(2-chloranyl-1,3-thiazol-5-yl)methyl]-9-methyl-pyrido[1,2-a]pyrimidine-2,4-dione, Acetylcholine-binding protein | | Authors: | Montgomery, M.G. | | Deposit date: | 2021-08-06 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural Biology-Guided Design, Synthesis, and Biological Evaluation of Novel Insect Nicotinic Acetylcholine Receptor Orthosteric Modulators.
J.Med.Chem., 65, 2022
|
|
8V36
 
 | |
7VMH
 
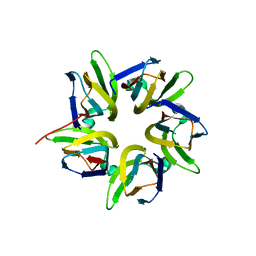 | |
7Z5X
 
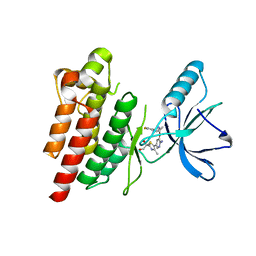 | | ROS1 with AstraZeneca ligand 2 | | Descriptor: | (2R)-2-[5-(6-amino-5-{(1R)-1-[2-(1,3-dihydro-2H-1,2,3-triazol-2-yl)-5-fluorophenyl]ethoxy}pyridin-3-yl)-4-methyl-1,3-thiazol-2-yl]propane-1,2-diol, Proto-oncogene tyrosine-protein kinase ROS | | Authors: | Hargreaves, D. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.035 Å) | | Cite: | Virtual Screening in the Cloud Identifies Potent and Selective ROS1 Kinase Inhibitors.
J.Chem.Inf.Model., 62, 2022
|
|
7N7Y
 
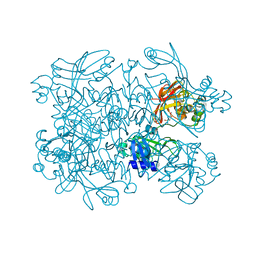 | | Crystal Structure of SARS-CoV-2 NendoU in complex with Z18197050 | | Descriptor: | Uridylate-specific endoribonuclease, methyl 4-sulfamoylbenzoate | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
7VWP
 
 | | Structure of the flavin-dependent monooxygenase FlsO1 from the biosynthesis of fluostatinsin | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FlsO1, PHOSPHATE ION, ... | | Authors: | Zhang, Y, Yang, C, Zhang, L, Zhang, C. | | Deposit date: | 2021-11-11 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural insights of multifunctional flavin-dependent monooxygenase FlsO1-catalyzed unexpected xanthone formation
Nat Commun, 13, 2022
|
|
5D91
 
 | | Structure of a phosphatidylinositolphosphate (PIP) synthase from Renibacterium Salmoninarum | | Descriptor: | AF2299 protein,Phosphatidylinositol synthase, MAGNESIUM ION, Octadecane, ... | | Authors: | Clarke, O.B, Tomasek, D.T, Jorge, C.D, Belcher Dufrisne, M, Kim, M, Banerjee, S, Rajashankar, K.R, Hendrickson, W.A, Santos, H, Mancia, F. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis for phosphatidylinositol-phosphate biosynthesis.
Nat Commun, 6, 2015
|
|
6MH9
 
 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein A121I mutant to 2.02 Angstrom resolution | | Descriptor: | Fatty Acid Kinase (Fak) B1 protein, PALMITIC ACID | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2018-09-17 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
