7ECV
 
 | | The Csy-AcrIF14 complex | | Descriptor: | AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L.X, Feng, Y. | | Deposit date: | 2021-03-13 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
7ELM
 
 | | Structure of Csy-AcrIF24 | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
7ELN
 
 | | Structure of Csy-AcrIF24-dsDNA | | Descriptor: | 54-MER DNA, AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
3ILK
 
 | | The structure of a probable methylase family protein from Haemophilus influenzae Rd KW20 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tan, K, Li, H, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The structure of a probable methylase family protein from Haemophilus influenzae Rd KW20
To be Published
|
|
7EU9
 
 | | Crystal structure of the selenomethionine(SeMet)-derived Cas12i1 R-loop complex before target DNA cleavage | | Descriptor: | CITRIC ACID, Cas12i1 D647A mutant, DNA (24-MER), ... | | Authors: | Zhang, B, Luo, D.Y, Li, Y, OuYang, S.Y. | | Deposit date: | 2021-05-16 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanistic insights into the R-loop formation and cleavage in CRISPR-Cas12i1.
Nat Commun, 12, 2021
|
|
2JLR
 
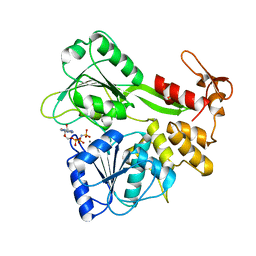 | | Dengue virus 4 NS3 helicase in complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SERINE PROTEASE SUBUNIT NS3 | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
5O69
 
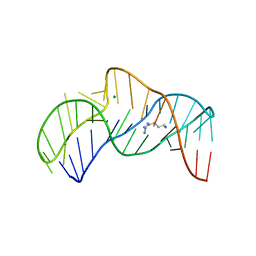 | | The structure of the thermobifida fusca guanidine III riboswitch with agmatine. | | Descriptor: | AGMATINE, MAGNESIUM ION, RNA (37-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-06-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
2JLS
 
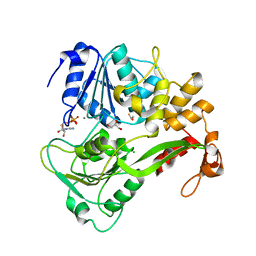 | | Dengue virus 4 NS3 helicase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
4NYZ
 
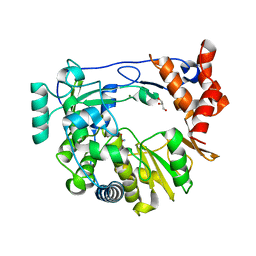 | | The EMCV 3Dpol structure with altered motif A conformation at 2.15A resolution | | Descriptor: | GLUTAMINE, GLYCEROL, Genome polyprotein, ... | | Authors: | Vives-adrian, L, Lujan, C, Ferrer-orta, C, Verdaguer, N. | | Deposit date: | 2013-12-11 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a cardiovirus RNA-dependent RNA polymerase reveals an unusual conformation of the polymerase active site
J.Virol., 88, 2014
|
|
4NZ0
 
 | |
5KTJ
 
 | | Crystal structure of Pistol, a class of self-cleaving ribozyme | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, Pistol (50-MER), ... | | Authors: | Nguyen, L.A, Wang, J, Steitz, T.A. | | Deposit date: | 2016-07-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structure of Pistol, a class of self-cleaving ribozyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3BO2
 
 | |
2JLQ
 
 | | Dengue virus 4 NS3 helicase structure, apo enzyme. | | Descriptor: | CHLORIDE ION, GLYCEROL, SERINE PROTEASE SUBUNIT NS3 | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
1M8Z
 
 | |
2RT3
 
 | | Solution structure of the second RRM domain of Nrd1 | | Descriptor: | Negative regulator of differentiation 1 | | Authors: | Kobayashi, A, Kanaba, T, Mishima, M. | | Deposit date: | 2013-04-16 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the second RRM domain of Nrd1, a fission yeast MAPK target RNA binding protein, and implication for its RNA recognition and regulation
Biochem.Biophys.Res.Commun., 437, 2013
|
|
1N9R
 
 | | Crystal structure of a heptameric ring complex of yeast SmF in spacegroup P4122 | | Descriptor: | Small nuclear ribonucleoprotein F | | Authors: | Collins, B.M, Cubeddu, L, Naidoo, N, Harrop, S.J, Kornfeld, G.D, Dawes, I.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2002-11-26 | | Release date: | 2002-12-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Homomeric ring assemblies of eukaryotic Sm proteins have affinity for
both RNA and DNA: Crystal structure of an oligomeric complex of yeast
SmF
J.Biol.Chem., 278, 2003
|
|
1WCL
 
 | |
5M84
 
 | | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSpA mRNA 5' cap analog | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-sulfanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanyl-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSpA mRNA 5' cap analog
To Be Published
|
|
5U0C
 
 | |
5M7Z
 
 | | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSepG mRNA 5' cap analog (beta-Se-ARCA D2) | | Descriptor: | CHLORIDE ION, Eukaryotic translation initiation factor 4E, GLYCEROL, ... | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSepG mRNA 5' cap analog (beta-Se-ARCA D2)
To Be Published
|
|
5M80
 
 | | Translation initiation factor 4E in complex with (RP)-iPr-m7GppSpG mRNA 5' cap analog | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [[[(3~{a}~{R},4~{R},6~{R},6~{a}~{R})-4-(2-azanyl-7-methyl-6-oxidanylidene-1~{H}-purin-7-ium-9-yl)-2,2-dimethyl-3~{a},4,6,6~{a}-tetrahydrofuro[3,4-d][1,3]dioxol-6-yl]methoxy-oxidanyl-phosphoryl]oxy-sulfanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Translation initiation factor 4E in complex with (RP)-iPr-m7GppSpG mRNA 5' cap analog
To Be Published
|
|
5QRH
 
 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury in complex with Z32327641 | | Descriptor: | CADMIUM ION, T-box transcription factor T, [4-(cyclopropanecarbonyl)piperazin-1-yl](furan-2-yl)methanone | | Authors: | Newman, J.A, Gavard, A.E, Fernandez-Cid, A, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QSL
 
 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury in complex with Z1273312153 | | Descriptor: | CADMIUM ION, N-methyl-1H-indole-7-carboxamide, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A.E, Fernandez-Cid, A, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5U0P
 
 | | Cryo-EM structure of the transcriptional Mediator | | Descriptor: | Mediator complex subunit 10, Mediator complex subunit 11, Mediator complex subunit 14, ... | | Authors: | Tsai, K.-L, Yu, X, Gopalan, S, Chao, T.-C, Zhang, Y, Florens, L, Washburn, M.P, Murakami, K, Conaway, R.C, Conaway, J.W, Asturias, F. | | Deposit date: | 2016-11-26 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mediator structure and rearrangements required for holoenzyme formation.
Nature, 544, 2017
|
|
4DWA
 
 | | Crystal structure of an active-site mutant of the glycoprotein Erns from the pestivirus BVDV-1 in complex with a CpUpC trinucleotide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, E(rns) glycoprotein, ... | | Authors: | Krey, T, Bontems, F, Vonrhein, C, Vaney, M.-C, Bricogne, G, Ruemenapf, T, Rey, F.A. | | Deposit date: | 2012-02-24 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structure of the Pestivirus Envelope Glycoprotein E(rns) and Mechanistic Analysis of Its Ribonuclease Activity.
Structure, 20, 2012
|
|
