6NFY
 
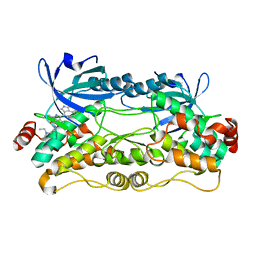 | | Crystal structure of nonphosphorylated, HPK1 kinase domain in complex with sunitinib in the inactive state. | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 1, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Johnson, E, McTigue, M, Cronin, C.N. | | Deposit date: | 2018-12-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Multiple conformational states of the HPK1 kinase domain in complex with sunitinib reveal the structural changes accompanying HPK1 trans-regulation.
J.Biol.Chem., 294, 2019
|
|
5WK3
 
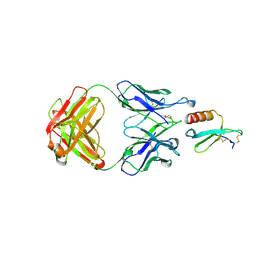 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN CCL17 AND M116 FAB | | Descriptor: | C-C motif chemokine 17, GLYCEROL, M116 HEAVY CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2017-07-24 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into chemokine CCL17 recognition by antibody M116.
Biochem Biophys Rep, 13, 2018
|
|
5WNM
 
 | |
5WK9
 
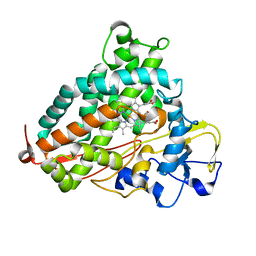 | | R186AP450cam with CN and camphor | | Descriptor: | CAMPHOR, CYANIDE ION, Camphor 5-monooxygenase, ... | | Authors: | Poulos, T.L, Batabyal, D. | | Deposit date: | 2017-07-24 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Effect of Redox Partner Binding on Cytochrome P450 Conformational Dynamics.
J. Am. Chem. Soc., 139, 2017
|
|
5WR9
 
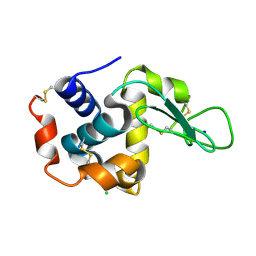 | | Crystal structure of hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Sugahara, M, Suzuki, M, Masuda, T, Inoue, S, Nango, E. | | Deposit date: | 2016-12-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
5X1R
 
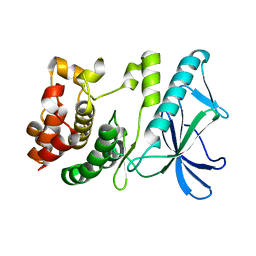 | | PpkA-294 apo form | | Descriptor: | PpkA-294 | | Authors: | Li, P.P, Ran, T.T, Xu, D.Q, Wang, W.W. | | Deposit date: | 2017-01-26 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the kinase domain of PpkA, a key regulatory component of T6SS, reveal a general inhibitory mechanism.
Biochem.J., 475, 2018
|
|
6NNY
 
 | |
6NWV
 
 | |
6NP6
 
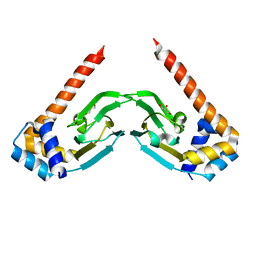 | | Crystal structure of the sensor domain of the transcriptional regulator HcpR from Porphyromonas Gingivalis | | Descriptor: | Crp/Fnr family transcriptional regulator, GLYCEROL | | Authors: | Musayev, F.N, Belvin, B.R, Escalante, C.R, Turner, J, Scarsdale, J.N, Lewis, J.P. | | Deposit date: | 2019-01-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nitrosative Stress Sensing in Porphyromonas gingivalis: Structure and Mechanisms of the Heme Binding Transcriptional Regulator HcpR.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6NUP
 
 | |
6NV7
 
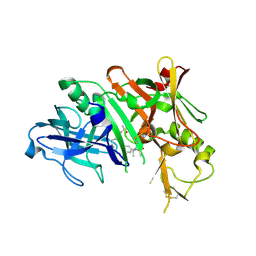 | | BACE1 in complex with a macrocyclic inhibitor | | Descriptor: | (E)-N-(2-methylpropylidene)-N~2~-{[(4S)-17-[(methylsulfonyl)(propyl)amino]-2-oxo-3-azatricyclo[13.3.1.1~6,10~]icosa-1(19),6(20),7,9,15,17-hexaen-4-yl]methyl}-D-threoninamide, Beta-secretase 1 | | Authors: | Yen, Y.C, Ghosh, A.K, Mesecar, A.D. | | Deposit date: | 2019-02-04 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.132 Å) | | Cite: | Development of an Efficient Enzyme Production and Structure-Based Discovery Platform for BACE1 Inhibitors.
Biochemistry, 58, 2019
|
|
6O21
 
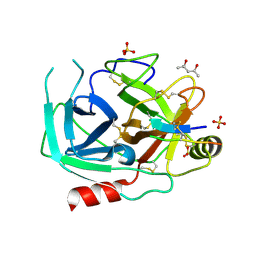 | | Crystal Structure of Human KLK4 in Complex With Cleaved SFTI-FCQR(Asn14)[1,14] Inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Kallikrein 4 (Prostase, enamel matrix, ... | | Authors: | Ilyichova, O.V, Buckle, A.M. | | Deposit date: | 2019-02-22 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | KLK4 Inhibition by Cyclic and Acyclic Peptides: Structural and Dynamical Insights into Standard-Mechanism Protease Inhibitors.
Biochemistry, 58, 2019
|
|
6NVB
 
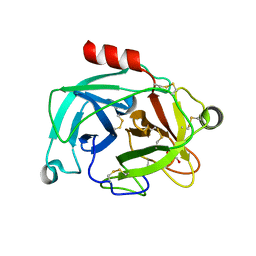 | | Crystal structure of the inhibitor-free form of the serine protease kallikrein-4 | | Descriptor: | GLYCEROL, Kallikrein-4, SULFATE ION | | Authors: | Riley, B.T, Buckle, A.M, McGowan, S. | | Deposit date: | 2019-02-04 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.636 Å) | | Cite: | Crystal structure of the inhibitor-free form of the serine protease kallikrein-4.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6O1S
 
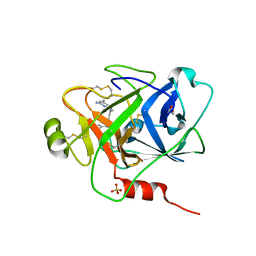 | | Structure of human plasma kallikrein protease domain with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-[(6-amino-2,4-dimethylpyridin-3-yl)methyl]-1-({4-[(1H-pyrazol-1-yl)methyl]phenyl}methyl)-1H-pyrazole-4-carboxamide, PHOSPHATE ION, ... | | Authors: | Partridge, J.R, Choy, R.M. | | Deposit date: | 2019-02-21 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of full-length plasma kallikrein bound to highly specific inhibitors describe a new mode of targeted inhibition.
J.Struct.Biol., 206, 2019
|
|
2IJ4
 
 | | Structure of the A264K mutant of cytochrome P450 BM3 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 | | Authors: | Toogood, H.S, Leys, D. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and spectroscopic characterization of P450 BM3 mutants with unprecedented P450 heme iron ligand sets. New heme ligation states influence conformational equilibria in P450 BM3.
J.Biol.Chem., 282, 2007
|
|
5EDB
 
 | |
2IKO
 
 | | Crystal Structure of Human Renin Complexed with Inhibitor | | Descriptor: | 5-{4-[(3,5-DIFLUOROBENZYL)AMINO]PHENYL}-6-ETHYLPYRIMIDINE-2,4-DIAMINE, Renin | | Authors: | Mochalkin, I. | | Deposit date: | 2006-10-02 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding thermodynamics of substituted diaminopyrimidine renin inhibitors.
Anal.Biochem., 360, 2007
|
|
5EH0
 
 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | DIMETHYL SULFOXIDE, Dual specificity protein kinase TTK, N2-(2-Methoxy-4-(1-methyl-1H-pyrazol-4-yl)phenyl)-N8-neopentylpyrido[3,4-d]pyrimidine-2,8-diamine | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-27 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle Kinase 1 (MPS1) Using a Structure-Based Hybridization Approach.
J.Med.Chem., 59, 2016
|
|
3EU1
 
 | | Crystal Structure determination of goat hemoglobin (Capra hircus) at 3 angstrom resolution | | Descriptor: | Hemoglobin subunit alpha-1/2, Hemoglobin subunit beta-A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sathya Moorthy, P, Neelagandan, K, Balasubramanian, M, Ponnuswamy, M.N. | | Deposit date: | 2008-10-09 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Purification, crystallization and preliminary X-ray diffraction studies on goat (Capra hircus) hemoglobin - a low oxygen affinity species
Protein Pept.Lett., 16, 2009
|
|
3E7Z
 
 | | Structure of human insulin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Timofeev, V.I, Lyashenko, A.V, Kislitsyn, Y.A, Baidus, A.N, Kuranova, I.P. | | Deposit date: | 2008-08-19 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human insulin
To be published
|
|
5EI0
 
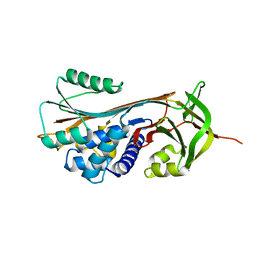 | | Structure of RCL-cleaved vaspin (serpinA12) | | Descriptor: | Serpin A12 | | Authors: | Pippel, J, Kuettner, B.E, Ulbricht, D, Daberger, J, Schultz, S, Heiker, J.T, Strater, N. | | Deposit date: | 2015-10-29 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of cleaved vaspin (serpinA12).
Biol.Chem., 397, 2016
|
|
2IOT
 
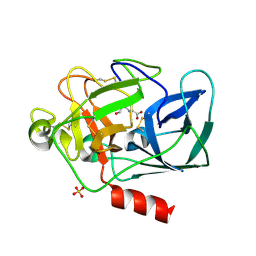 | | Clavulanic Acid bound to Elastase | | Descriptor: | Elastase-1, N-(3-OXOPROPYL)GLYCINE, SULFATE ION | | Authors: | Farady, C, Navia, M.A. | | Deposit date: | 2006-10-10 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elastase Inhibition by Clavulanic Acid, and Inhibitor of Bacterial b-lactamases: Mechanistic and Structural Studies
To be Published
|
|
6ZHI
 
 | |
2IT6
 
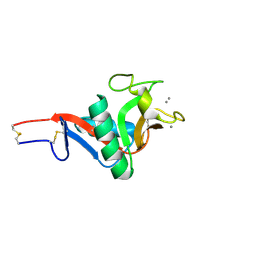 | | Crystal Structure of DCSIGN-CRD with man2 | | Descriptor: | CALCIUM ION, CD209 antigen, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Weis, W.I, Feinberg, H, Castelli, R, Drickamer, K, Seeberger, P.H. | | Deposit date: | 2006-10-19 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Multiple modes of binding enhance the affinity of DC-SIGN for high mannose N-linked glycans found on viral glycoproteins.
J.Biol.Chem., 282, 2007
|
|
3EBZ
 
 | | High Resolution HIV-2 Protease Structure in Complex with Clinical Drug Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2008-08-28 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural evidence for effectiveness of darunavir and two related antiviral inhibitors against HIV-2 protease
J.Mol.Biol., 384, 2008
|
|
