3FDE
 
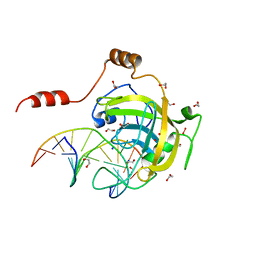 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG DNA, crystal structure in space group C222(1) at 1.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', ... | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-25 | | Release date: | 2009-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
2NXW
 
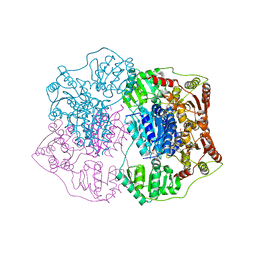 | | Crystal structure of phenylpyruvate decarboxylase of Azospirillum brasilense | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Versees, W, Spaepen, S, Vanderleyden, J, Steyaert, J. | | Deposit date: | 2006-11-20 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of phenylpyruvate decarboxylase from Azospirillum brasilense at 1.5 A resolution. Implications for its catalytic and regulatory mechanism.
Febs J., 274, 2007
|
|
1W9S
 
 | |
4FXF
 
 | |
2GLR
 
 | |
4GFP
 
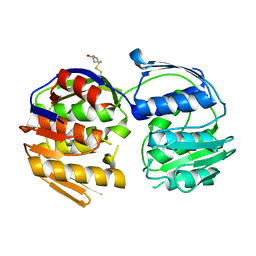 | | 2.7 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in a second conformational state | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, BETA-MERCAPTOETHANOL | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2.7 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in second conformational state
TO BE PUBLISHED
|
|
3WDW
 
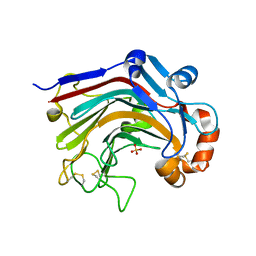 | | The apo-form structure of E113A from Paecilomyces thermophila | | Descriptor: | Beta-1,3-1,4-glucanase, SULFATE ION | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
1Y2H
 
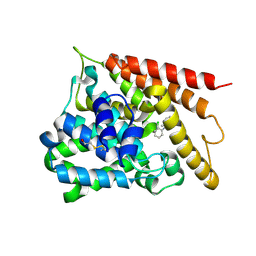 | | Catalytic Domain Of Human Phosphodiesterase 4B In Complex With 1-(2-chloro-phenyl)-3,5-dimethyl-1H-pyrazole-4-carboxylic acid ethyl ester | | Descriptor: | 1-(2-CHLOROPHENYL)-3,5-DIMETHYL-1H-PYRAZOLE-4-CARBOXYLIC ACID ETHYL ESTER, MAGNESIUM ION, ZINC ION, ... | | Authors: | Card, G.L, Blasdel, L, England, B.P, Zhang, C, Suzuki, Y, Gillette, S, Fong, D, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design
Nat.Biotechnol., 23, 2005
|
|
3NON
 
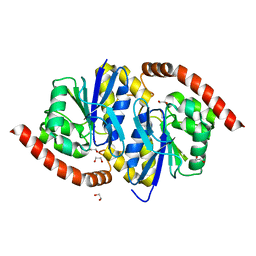 | | Crystal Structure of Isocyanide Hydratase from Pseudomonas fluorescens | | Descriptor: | 1,2-ETHANEDIOL, Isocyanide hydratase | | Authors: | Lakshminarasimhan, M, Madzelan, P, Nan, R, Milkovic, N.M, Wilson, M.A. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Evolution of New Enzymatic Function by Structural Modulation of Cysteine Reactivity in Pseudomonas fluorescens Isocyanide Hydratase.
J.Biol.Chem., 285, 2010
|
|
1GAT
 
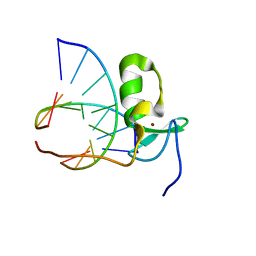 | | SOLUTION STRUCTURE OF THE SPECIFIC DNA COMPLEX OF THE ZINC CONTAINING DNA BINDING DOMAIN OF THE ERYTHROID TRANSCRIPTION FACTOR GATA-1 BY MULTIDIMENSIONAL NMR | | Descriptor: | DNA (5'-D(P*AP*GP*AP*TP*AP*AP*AP*C)3'), DNA (5'-D(P*GP*TP*TP*TP*AP*TP*CP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | Authors: | Clore, G.M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1993-06-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1.
Science, 261, 1993
|
|
1GAU
 
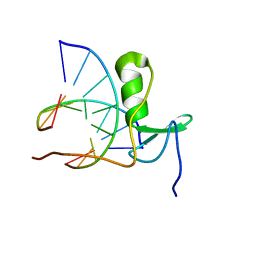 | |
4ISN
 
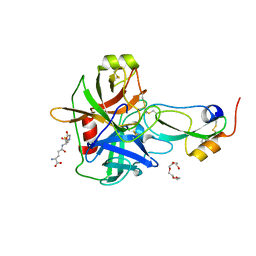 | | Crystal Structure of Matriptase in complex with its inhibitor HAI-1 | | Descriptor: | GLUTATHIONE, Kunitz-type protease inhibitor 1, Suppressor of tumorigenicity 14 protein, ... | | Authors: | Huang, M.D, Zhao, B.Y, Yuan, C, Li, R. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of matriptase in complex with its inhibitor hepatocyte growth factor activator inhibitor-1.
J.Biol.Chem., 288, 2013
|
|
4FZ9
 
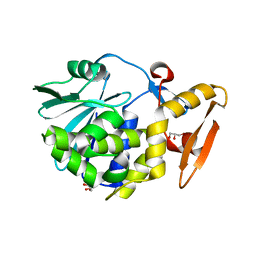 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica Balsamina with disaccharide, N-Acetylglucosamine (beta-1, 4) Mannose at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-07-06 | | Release date: | 2012-08-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the complex of Ribosome inactivating protein from Momordica Balsamina with disaccharide, N-Acetylglucosamine (beta-1, 4) Mannose at 1.7 A resolution
To be Published
|
|
1GPS
 
 | | SOLUTION STRUCTURE OF GAMMA 1-H AND GAMMA 1-P THIONINS FROM BARLEY AND WHEAT ENDOSPERM DETERMINED BY 1H-NMR: A STRUCTURAL MOTIF COMMON TO TOXIC ARTHROPOD PROTEINS | | Descriptor: | GAMMA-1-P THIONIN | | Authors: | Bruix, M, Jimenez, M.A, Santoro, J, Gonzalez, C, Colilla, F.J, Mendez, E, Rico, M. | | Deposit date: | 1992-07-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of gamma 1-H and gamma 1-P thionins from barley and wheat endosperm determined by 1H-NMR: a structural motif common to toxic arthropod proteins.
Biochemistry, 32, 1993
|
|
4G5F
 
 | | Pantothenate synthetase in complex with racemate (2S)-2,3-DIHYDRO-1,4-BENZODIOXINE-2-CARBOXYLIC ACID and (2R)-2,3-DIHYDRO-1,4-BENZODIOXINE-2-CARBOXYLIC ACID | | Descriptor: | (2R)-2,3-dihydro-1,4-benzodioxine-2-carboxylic acid, (2S)-2,3-dihydro-1,4-benzodioxine-2-carboxylic acid, 1,2-ETHANEDIOL, ... | | Authors: | Silvestre, H.L, Blundell, T.L, Abell, C, Ciulli, A. | | Deposit date: | 2012-07-17 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Integrated biophysical approach to fragment screening and validation for fragment-based lead discovery.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2P2N
 
 | | Crystal Structure and Allosteric Regulation of the Cytoplasmic Escherichia coli L-Asparaginase I | | Descriptor: | 1,2-ETHANEDIOL, ASPARAGINE, ASPARTIC ACID, ... | | Authors: | Yun, M.-K, Nourse, A, White, S.W, Rock, C.O, Heath, R.J. | | Deposit date: | 2007-03-07 | | Release date: | 2007-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Allosteric Regulation of the Cytoplasmic Escherichia colil-Asparaginase I
J.Mol.Biol., 369, 2007
|
|
3TSS
 
 | | TOXIC SHOCK SYNDROME TOXIN-1 TETRAMUTANT, P2(1) CRYSTAL FORM | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Prasad, G.S, Radhakrishnan, R, Mitchell, D.T, Earhart, C.A, Dinges, M.M, Cook, W.J, Schlivert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-12-11 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined structures of three crystal forms of toxic shock syndrome toxin-1 and of a tetramutant with reduced activity.
Protein Sci., 6, 1997
|
|
3QQU
 
 | | Cocrystal structure of unphosphorylated igf with pyrimidine 8 | | Descriptor: | Insulin-like growth factor 1 receptor, N~2~-[3-methoxy-4-(morpholin-4-yl)phenyl]-N~4~-(quinolin-3-yl)pyrimidine-2,4-diamine | | Authors: | Huang, X. | | Deposit date: | 2011-02-16 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of 2,4-bis-arylamino-1,3-pyrimidines as insulin-like growth factor-1 receptor (IGF-1R) inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1HJC
 
 | |
1HM0
 
 | | CRYSTAL STRUCTURE OF S.PNEUMONIAE N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE, GLMU | | Descriptor: | CALCIUM ION, N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE | | Authors: | Sulzenbacher, G, Gal, L, Peneff, C, Fassy, F, Bourne, Y. | | Deposit date: | 2000-12-04 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase bound to acetyl-coenzyme A reveals a novel active site architecture.
J.Biol.Chem., 276, 2001
|
|
4GX3
 
 | |
4L15
 
 | | Crystal structure of FIGL-1 AAA domain | | Descriptor: | Fidgetin-like protein 1, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Peng, W, Lin, Z, Li, W, Lu, J, Shen, Y, Wang, C. | | Deposit date: | 2013-06-02 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the unusually strong ATPase activity of the AAA domain of the Caenorhabditis elegans fidgetin-like 1 (FIGL-1) protein.
J.Biol.Chem., 288, 2013
|
|
4N6P
 
 | | Crystal Structure of C-lobe of Bovine lactoferrin complexed with meclofenamic acid at 1.4 A resolution | | Descriptor: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gautam, L, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-10-14 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structre of C-LOBE OF BOVINE LACTOFERRIN COMPLEXED WITH MECLOFENAMIC ACID AT 1.4 A RESOLUTION
TO BE PUBLISHED
|
|
1E0W
 
 | | Xylanase 10A from Sreptomyces lividans. native structure at 1.2 angstrom resolution | | Descriptor: | ENDO-1,4-BETA-XYLANASE A | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
3EOC
 
 | | Cdk2/CyclinA complexed with a imidazo triazin-2-amine | | Descriptor: | 5-methyl-7-phenyl-N-(3,4,5-trimethoxyphenyl)imidazo[5,1-f][1,2,4]triazin-2-amine, Cell division protein kinase 2, Cyclin-A2 | | Authors: | Cheung, M, Kuntz, K, Pobanz, M, Salovich, J, Wilson, B, Andrews, W, Shewchuk, L, Epperly, A, Hassler, D, Leesnitzer, M, Smith, J, Smith, G, Lansing, T, Mook, R. | | Deposit date: | 2008-09-26 | | Release date: | 2008-11-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Imidazo[5,1-f][1,2,4]triazin-2-amines as novel inhibitors of polo-like kinase 1.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
