3NAX
 
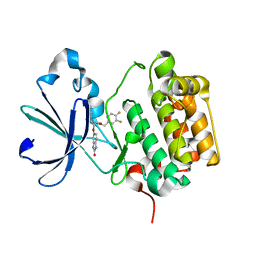 | | PDK1 in complex with inhibitor MP7 | | Descriptor: | 1-(3,4-difluorobenzyl)-2-oxo-N-{(1R)-2-[(2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)oxy]-1-phenylethyl}-1,2-dihydropyridine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1 | | Authors: | Yan, Y, Munshi, S.K, Allison, T. | | Deposit date: | 2010-06-02 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Genetic and pharmacological inhibition of PDK1 in cancer cells: characterization of a selective allosteric kinase inhibitor.
J.Biol.Chem., 286, 2011
|
|
3CTB
 
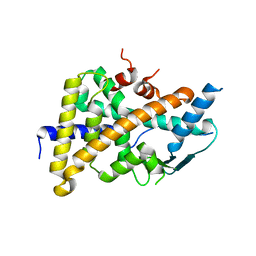 | | Tethered PXR-LBD/SRC-1p apoprotein | | Descriptor: | Pregnane X receptor, Linker, Steroid receptor coactivator 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2008-04-11 | | Release date: | 2008-12-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Construction and characterization of a fully active PXR/SRC-1 tethered protein with increased stability
Protein Eng.Des.Sel., 21, 2008
|
|
1DJX
 
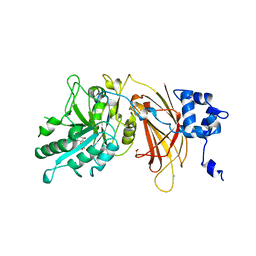 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-1,4,5-TRISPHOSPHATE | | Descriptor: | ACETATE ION, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
3NBQ
 
 | |
3CKR
 
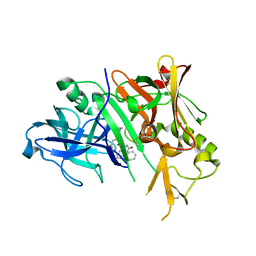 | | Crystal structure of BACE-1 in complex with inhibitor | | Descriptor: | (4S)-1,4-dibenzyl-N-[(1S,2R)-1-benzyl-3-{[3-(dimethylamino)benzyl]amino}-2-hydroxypropyl]-2-oxoimidazolidine-4-carboxamide, Beta-secretase 1 | | Authors: | Min, K. | | Deposit date: | 2008-03-16 | | Release date: | 2008-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthesis, SAR, and X-ray structure of human BACE-1 inhibitors with cyclic urea derivatives
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3N6K
 
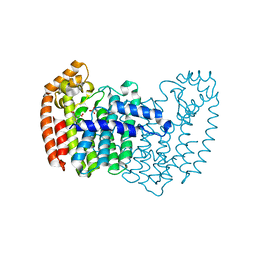 | | Human FPPS complex with NOV_823 | | Descriptor: | 1-(carboxymethyl)-1H-benzo[g]indole-2-carboxylic acid, FARNESYL PYROPHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Rondeau, J.-M. | | Deposit date: | 2010-05-26 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Allosteric non-bisphosphonate FPPS inhibitors identified by fragment-based discovery.
Nat.Chem.Biol., 6, 2010
|
|
3N84
 
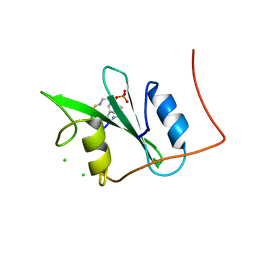 | |
3N8S
 
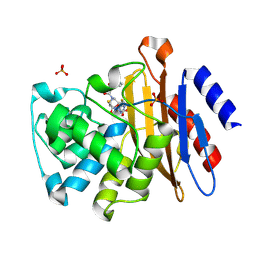 | | Crystal Structure of BlaC-E166A covalently bound with Cefamandole | | Descriptor: | (2R)-2-[(1R)-1-{[(2R)-2-hydroxy-2-phenylacetyl]amino}-2-oxoethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W, Blanchard, J.S. | | Deposit date: | 2010-05-28 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Michaelis Complex (1.2 A) and the Covalent Acyl Intermediate (2.0 A) of Cefamandole Bound in the Active Sites of the Mycobacterium tuberculosis beta-Lactamase K73A and E166A Mutants.
Biochemistry, 49, 2010
|
|
1J5J
 
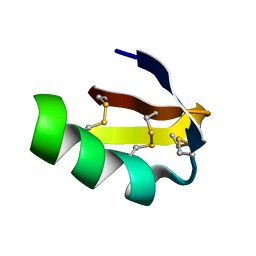 | | Solution structure of HERG-specific scorpion toxin BeKm-1 | | Descriptor: | BeKm-1 toxin | | Authors: | Korolokova, Y.V, Bocharov, E.V, Angelo, K, Maslennikov, I.V, Grinenko, O.V, Lipkin, A.V, Nosireva, E.D, Pluzhnikov, K.A, Olesen, S.-P, Arseniev, A.S, Grishin, E.V. | | Deposit date: | 2002-04-16 | | Release date: | 2002-11-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | New binding site on common molecular scaffold provides HERG channel specificity of scorpion toxin BeKm-1.
J.Biol.Chem., 277, 2002
|
|
3TNN
 
 | |
1INV
 
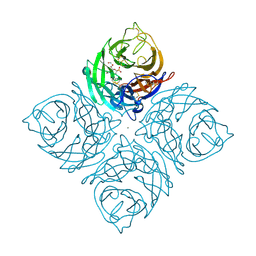 | | A SIALIC ACID DERIVED PHOSPHONATE ANALOG INHIBITS DIFFERENT STRAINS OF INFLUENZA VIRUS NEURAMINIDASE WITH DIFFERENT EFFICIENCIES | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | White, C.L, Janakiraman, M.N, Laver, W.G, Philippon, C, Vasella, A, Air, G.M, Luo, M. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A sialic acid-derived phosphonate analog inhibits different strains of influenza virus neuraminidase with different efficiencies.
J.Mol.Biol., 245, 1995
|
|
4EQI
 
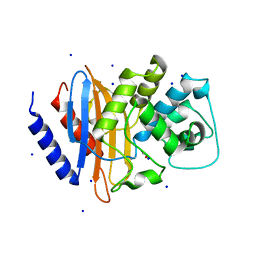 | | Crystal structure of serratia fonticola carbapenemase SFC-1 | | Descriptor: | 1,2-ETHANEDIOL, Carbapenem-hydrolizing beta-lactamase SFC-1, SODIUM ION | | Authors: | Fonseca, F, Spencer, J. | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The basis for carbapenem hydrolysis by class A beta-lactamases: a combined investigation using crystallography and simulations.
J.Am.Chem.Soc., 134, 2012
|
|
1EBH
 
 | |
2O2D
 
 | | Crystal structure of phosphoglucose isomerase from Trypanosoma brucei complexed with citrate | | Descriptor: | CITRIC ACID, GLYCEROL, Glucose-6-phosphate isomerase, ... | | Authors: | Arsenieva, D, Mazock, G.H, Appavu, B.L, Jeffery, C.J. | | Deposit date: | 2006-11-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of phosphoglucose isomerase from Trypanosoma brucei complexed with glucose-6-phosphate at 1.6 A resolution
Proteins, 74, 2008
|
|
2ESX
 
 | | The structure of the V3 region within gp120 of JR-FL HIV-1 strain (minimized average structure) | | Descriptor: | Envelope polyprotein GP160 | | Authors: | Rosen, O, Sharon, M, Samson, A.O, Quadt, S.R, Anglister, J. | | Deposit date: | 2005-10-27 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular switch for alternative conformations of the HIV-1 V3 region: Implications for phenotype conversion.
Proc.Natl.Acad.Sci.Usa, 13, 2006
|
|
2ESZ
 
 | | The structure of the V3 region within gp120 of JR-FL HIV-1 strain (ensemble) | | Descriptor: | Envelope polyprotein GP160 | | Authors: | Rosen, O, Sharon, M, Samson, A.O, Quadt, S.R, Anglister, J. | | Deposit date: | 2005-10-27 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular switch for alternative conformations of the HIV-1 V3 region: Implications for phenotype conversion.
Proc.Natl.Acad.Sci.Usa, 13, 2006
|
|
1XM2
 
 | | Crystal structure of Human PRL-1 | | Descriptor: | SULFATE ION, Tyrosine Phosphatase | | Authors: | Jeong, D.G, Kim, S.J, Kim, J.H, Son, J.H, Ryu, S.E. | | Deposit date: | 2004-10-01 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Trimeric structure of PRL-1 phosphatase reveals an active enzyme conformation and regulation mechanisms
J.Mol.Biol., 345, 2005
|
|
3BCS
 
 | | Glycogen Phosphorylase complex with 1(-D-glucopyranosyl) uracil | | Descriptor: | 1-beta-D-glucopyranosylpyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Sovantzis, D.A, Hadjiloi, T, Hayes, J.M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G. | | Deposit date: | 2007-11-13 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D-Glucopyranosyl pyrimidine nucleoside binding to muscle glycogen phosphorylase b
To be Published
|
|
4L41
 
 | |
3TKU
 
 | | MRCK beta in complex with fasudil | | Descriptor: | 1,2-ETHANEDIOL, 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, Serine/threonine-protein kinase MRCK beta | | Authors: | Heikkila, T.J, Turnbull, A, Wheatley, E, Schroder, E, Crighton, D, Olson, M.F. | | Deposit date: | 2011-08-29 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Co-crystal structures of inhibitors with MRCK, a key regulator of tumor cell invasion
Plos One, 6, 2011
|
|
3BJF
 
 | | Pyruvate kinase M2 is a phosphotyrosine binding protein | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Wu, N. | | Deposit date: | 2007-12-03 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Pyruvate kinase M2 is a phosphotyrosine-binding protein.
Nature, 452, 2008
|
|
4PS3
 
 | |
1G3L
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TDP-L-RHAMNOSE COMPLEX. | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
2FCY
 
 | | HIV-1 DIS kissing-loop in complex with Neomycin | | Descriptor: | CHLORIDE ION, HIV-1 DIS RNA, NEOMYCIN, ... | | Authors: | Ennifar, E, Paillart, J.C, Marquet, R, Dumas, P. | | Deposit date: | 2005-12-13 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting the dimerization initiation site of HIV-1 RNA with aminoglycosides: from crystal to cell.
Nucleic Acids Res., 34, 2006
|
|
3RL9
 
 | | Crystal Structure of the complex of type I RIP with the hydrolyzed product of dATP, adenine at 1.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-04-19 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states
Biochim.Biophys.Acta, 1824, 2012
|
|
