8DXN
 
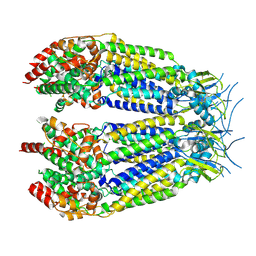 | | Structure of LRRC8C-LRRC8A(IL125) Chimera, Class 1 | | Descriptor: | Volume-regulated anion channel subunit LRRC8C,Volume-regulated anion channel subunit LRRC8A | | Authors: | Takahashi, H, Yamada, T, Denton, J.S, Strange, K, Karakas, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of a LRRC8 chimera with native functional properties reveal heptameric assembly.
Elife, 12, 2023
|
|
6UDY
 
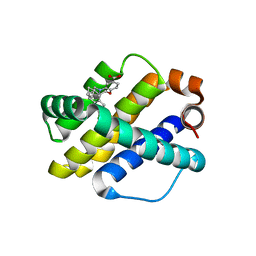 | | X-ray co-crystal structure of compound 5 with Mcl-1 | | Descriptor: | (3S)-6'-chloro-5-(cyclobutylmethyl)-3',4,4',5-tetrahydro-2H,2'H-spiro[1,5-benzoxazepine-3,1'-naphthalene]-7-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-09-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and in Vivo Evaluation of Macrocyclic Mcl-1 Inhibitors Featuring an alpha-Hydroxy Phenylacetic Acid Pharmacophore or Bioisostere.
J.Med.Chem., 62, 2019
|
|
6QPI
 
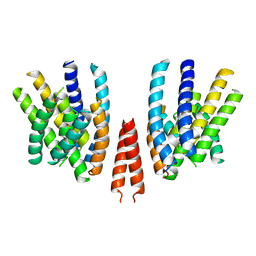 | | Cryo-EM structure of calcium-free mTMEM16F lipid scramblase in nanodisc | | Descriptor: | Anoctamin-6 | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
8E7X
 
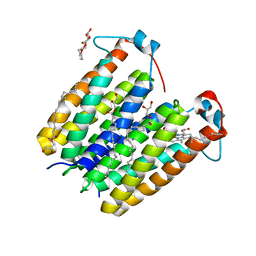 | | RsTSPO A138F with one Heme bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan-rich sensory protein | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
6QVC
 
 | |
8E7Y
 
 | | RsTSPO A138F with two heme bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan-rich sensory protein | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
6TXQ
 
 | | The high resolution structure of the FERM domain and helical linker of human moesin | | Descriptor: | ACETATE ION, Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Kelly, J.J, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
2LEG
 
 | | Membrane protein complex DsbB-DsbA structure by joint calculations with solid-state NMR and X-ray experimental data | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein DsbA, UBIQUINONE-1, ... | | Authors: | Tang, M, Sperling, L.J, Berthold, D.A, Schwieters, C.D, Nesbitt, A.E, Nieuwkoop, A.J, Gennis, R.B, Rienstra, C.M. | | Deposit date: | 2011-06-15 | | Release date: | 2011-10-26 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution membrane protein structure by joint calculations with solid-state NMR and X-ray experimental data.
J.Biomol.Nmr, 51, 2011
|
|
6QRM
 
 | | HsNMT1 in complex with both MyrCoA and GNCFSKRRAA substrates | | Descriptor: | Apoptosis-inducing factor 3, CHLORIDE ION, COENZYME A, ... | | Authors: | Dian, C, Riviere, F.B, Asensio, T, Giglione, C, Meinnel, T. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
8E7Z
 
 | | RsTSPO mutant -A138F | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Tryptophan-rich sensory protein | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
6UIR
 
 | | HIV-1 M184V reverse transcriptase-DNA complex with (-)-FTC-TP | | Descriptor: | MAGNESIUM ION, Primer DNA, SULFATE ION, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2019-10-01 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.63900113 Å) | | Cite: | Elucidating molecular interactions ofL-nucleotides with HIV-1 reverse transcriptase and mechanism of M184V-caused drug resistance.
Commun Biol, 2, 2019
|
|
4CDT
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-((3-Fluorophenethylamino)ethyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[2-[2-(3-fluorophenyl)ethylamino]ethyl]quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-11-05 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Simplified 2-Aminoquinoline-Based Scaffold for Potent and Selective Neuronal Nitric Oxide Synthase Inhibition.
J.Med.Chem., 57, 2014
|
|
8E7W
 
 | | RsTSPO A139T with Heme | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
6UJT
 
 | | P-glycoprotein mutant-Y303A and C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
8DMY
 
 | | Cryo-EM structure of cardiac muscle alpha-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha cardiac muscle 1, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-09 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
6U1U
 
 | | Human Angiopoietin-Like 4 C-Terminal Domain (cANGPTL4) with Palmitic Acid | | Descriptor: | Angiopoietin-related protein 4, PALMITIC ACID | | Authors: | Tarver, C.L, Yuan, Q, Singhal, A.J, Ramaker, R, Cooper, S, Pusey, M.L. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Angiopoietin-Like 4 C-Terminal Domain (cANGPTL4) Reveal a Binding Pocket with Multiple Ligands
To Be Published
|
|
4C7R
 
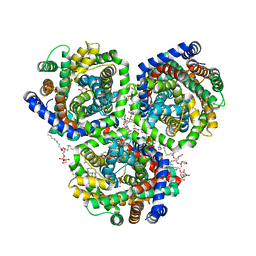 | | Inward facing conformation of the trimeric betaine transporter BetP in complex with lipids | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, CHLORIDE ION, ... | | Authors: | Koshy, C, Yildiz, O, Ziegler, C. | | Deposit date: | 2013-09-24 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Evidence for Functional Lipid Interactions in the Betaine Transporter Betp
Embo J., 32, 2013
|
|
8DNF
 
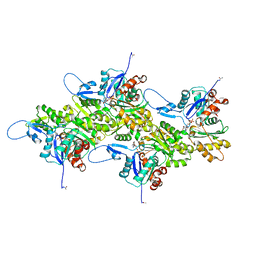 | | Cryo-EM structure of nonmuscle gamma-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 2, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
8DMX
 
 | | Cryo-EM structure of skeletal muscle alpha-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
8DNH
 
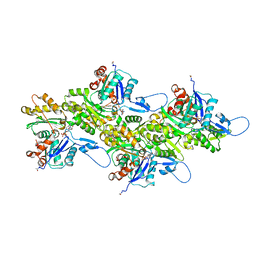 | | Cryo-EM structure of nonmuscle beta-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Arora, A.S, Huang, H.L, Heissler, S.M, Chinthalapudi, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural insights into actin isoforms.
Elife, 12, 2023
|
|
6QVO
 
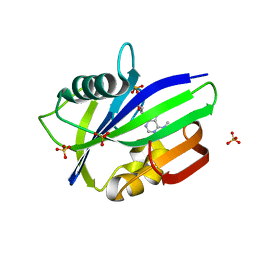 | | Crystal structure of human MTH1 in complex with N6-methyl-dAMP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, N6-METHYL-DEOXY-ADENOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Scaletti, E, Vallin, K.S, Brautigam, L, Sarno, A, Warpman Berglund, U, Helleday, T, Stenmark, P, Jemth, A.S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | MutT homologue 1 (MTH1) removes N6-methyl-dATP from the dNTP pool.
J.Biol.Chem., 295, 2020
|
|
8E6L
 
 | | X-ray structure of the Deinococcus radiodurans Nramp/MntH divalent transition metal transporter D296A mutant in an inward-open, manganese-bound state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Divalent metal cation transporter MntH, ... | | Authors: | Ray, S, Gaudet, R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | High-resolution structures with bound Mn 2+ and Cd 2+ map the metal import pathway in an Nramp transporter.
Elife, 12, 2023
|
|
6QZ9
 
 | | The cryo-EM structure of the collar complex and tail axis in bacteriophage phi29 | | Descriptor: | Portal protein, Pre-neck appendage protein, Proximal tail tube connector protein | | Authors: | Xu, J, Wang, D, Gui, M, Xiang, Y. | | Deposit date: | 2019-03-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural assembly of the tailed bacteriophage φ29.
Nat Commun, 10, 2019
|
|
6QLP
 
 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative 3 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, CHLORIDE ION, Galectin-3, ... | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.081 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
6QLU
 
 | | Galectin-3C in complex with fluoroaryltriazole monothiogalactoside derivative-8 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-(4-fluoranyl-3-methyl-phenyl)-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
