1RCQ
 
 | | The 1.45 A crystal structure of alanine racemase from a pathogenic bacterium, Pseudomonas aeruginosa, contains both internal and external aldimine forms | | Descriptor: | D-LYSINE, PYRIDOXAL-5'-PHOSPHATE, catabolic alanine racemase DadX | | Authors: | Le Magueres, P, Im, H, Dvorak, A, Strych, U, Benedik, M, Krause, K.L. | | Deposit date: | 2003-11-04 | | Release date: | 2004-06-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure at 1.45 A resolution of alanine racemase from a pathogenic bacterium, Pseudomonas aeruginosa, contains both internal and external aldimine forms.
Biochemistry, 42, 2003
|
|
1JK8
 
 | |
5JEA
 
 | | Structure of a cytoplasmic 11-subunit RNA exosome complex including Ski7, bound to RNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Exosome complex component CSL4, Exosome complex component MTR3, ... | | Authors: | Kowalinski, E, Ebert, J, Stegmann, E, Conti, E. | | Deposit date: | 2016-04-18 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of a Cytoplasmic 11-Subunit RNA Exosome Complex.
Mol.Cell, 63, 2016
|
|
5JJJ
 
 | | Structure of the SRII/HtrII Complex in P64 space group ("U" shape) | | Descriptor: | EICOSANE, RETINAL, Sensory rhodopsin II transducer, ... | | Authors: | Ishchenko, A, Round, E, Borshchevskiy, V, Grudinin, S, Gushchin, I, Klare, J, Remeeva, A, Polovinkin, V, Utrobin, P, Balandin, T, Engelhard, M, Bueldt, G, Gordeliy, V. | | Deposit date: | 2016-04-24 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New Insights on Signal Propagation by Sensory Rhodopsin II/Transducer Complex.
Sci Rep, 7, 2017
|
|
1R1T
 
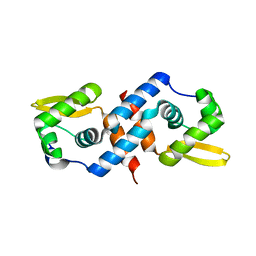 | | Crystal structure of the cyanobacterial metallothionein repressor SmtB in the apo-form | | Descriptor: | Transcriptional repressor smtB | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
5JJN
 
 | | Structure of the SRII/HtrII(G83F) Complex in P212121 space group ("V" shape) | | Descriptor: | EICOSANE, RETINAL, Sensory rhodopsin II transducer, ... | | Authors: | Ishchenko, A, Round, E, Borshchevskiy, V, Grudinin, S, Gushchin, I, Klare, J, Remeeva, A, Polovinkin, V, Utrobin, P, Balandin, T, Engelhard, M, Bueldt, G, Gordeliy, V. | | Deposit date: | 2016-04-24 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | New Insights on Signal Propagation by Sensory Rhodopsin II/Transducer Complex.
Sci Rep, 7, 2017
|
|
1LOQ
 
 | |
5JJV
 
 | |
1LP6
 
 | |
1RB8
 
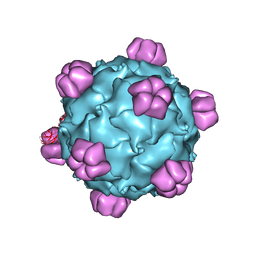 | | The phiX174 DNA binding protein J in two different capsid environments. | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, Capsid protein, DNA (5'-D(P*CP*AP*AP*A)-3'), ... | | Authors: | Bernal, R.A, Hafenstein, S, Esmeralda, R, Fane, B.A, Rossmann, M.G. | | Deposit date: | 2003-11-03 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The phiX174 Protein J Mediates DNA Packaging and Viral Attachment to Host Cells.
J.Mol.Biol., 337, 2004
|
|
1L2Q
 
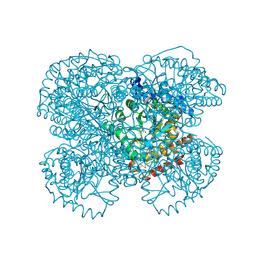 | | Crystal Structure of the Methanosarcina barkeri Monomethylamine Methyltransferase (MtmB) | | Descriptor: | AMMONIUM ION, monomethylamine methyltransferase | | Authors: | Hao, B, Gong, W, Ferguson, T.K, James, C.M, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2002-02-24 | | Release date: | 2002-06-05 | | Last modified: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A new UAG-encoded residue in the structure of a methanogen methyltransferase.
Science, 296, 2002
|
|
1RF0
 
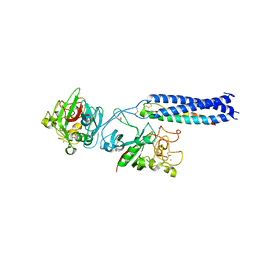 | | Crystal Structure of Fragment D of gammaE132A Fibrinogen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fibrinogen alpha/alpha-E chain, ... | | Authors: | Kostelansky, M.S, Gorkun, O.V, Lord, S.T. | | Deposit date: | 2003-11-07 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Calcium-Binding Site beta2, Adjacent to the "b" Polymerization Site, Modulates Lateral Aggregation of Protofibrils during Fibrin Polymerization.
Biochemistry, 43, 2004
|
|
1LE8
 
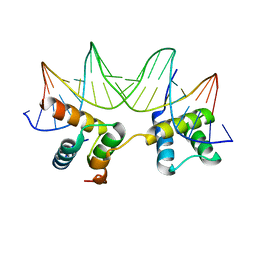 | | Crystal Structure of the MATa1/MATalpha2-3A Heterodimer Bound to DNA Complex | | Descriptor: | 5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*AP*AP*AP*TP*TP*TP*AP*CP*AP*TP*CP*A)-3', 5'-D(*TP*TP*GP*AP*TP*GP*TP*AP*AP*AP*TP*TP*TP*TP*TP*AP*CP*AP*TP*G)-3', MATING-TYPE PROTEIN A-1, ... | | Authors: | Ke, A, Mathias, J.R, Vershon, A.K, Wolberger, C. | | Deposit date: | 2002-04-09 | | Release date: | 2002-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Thermodynamic Characterization of the DNA Binding Properties of a Triple Alanine Mutant of MATalpha2
Structure, 10, 2002
|
|
5L5Q
 
 | |
5L66
 
 | |
1KIX
 
 | |
5L5O
 
 | | Yeast 20S proteasome with human beta5i (1-138) and human beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 16 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
5L65
 
 | |
1KLY
 
 | |
1KM4
 
 | |
1KGD
 
 | | Crystal Structure of the Guanylate Kinase-like Domain of Human CASK | | Descriptor: | FORMIC ACID, PERIPHERAL PLASMA MEMBRANE CASK | | Authors: | Li, Y, Spangenberg, O, Paarmann, I, Konrad, M, Lavie, A. | | Deposit date: | 2001-11-26 | | Release date: | 2001-12-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.314 Å) | | Cite: | Structural basis for nucleotide-dependent regulation of membrane-associated guanylate kinase-like domains.
J.Biol.Chem., 277, 2002
|
|
1KXP
 
 | |
1KHT
 
 | |
1KNM
 
 | | Streptomyces lividans Xylan Binding Domain cbm13 in Complex with Lactose | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Notenboom, V, Boraston, A.B, Williams, S.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-12-19 | | Release date: | 2002-06-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding.
Biochemistry, 41, 2002
|
|
1KC8
 
 | | Co-crystal Structure of Blasticidin S Bound to the 50S Ribosomal Subunit | | Descriptor: | 23S RRNA, 5S RRNA, BLASTICIDIN S, ... | | Authors: | Hansen, J.L, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-11-07 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structures of Five Antibiotics Bound at the Peptidyl Transferase Center of
the Large Ribosomal Subunit
J.Mol.Biol., 330, 2003
|
|
