5HP4
 
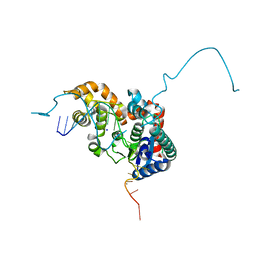 | | Crystal structure bacteriohage T5 D15 flap endonuclease (D155K) pseudo-enzyme-product complex with DNA and metal ions | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*AP*TP*CP*TP*AP*TP*AP*TP*GP*CP*CP*AP*TP*CP*GP*G)-3'), Exodeoxyribonuclease, ... | | Authors: | Almalki, F.A, Zhang, J, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
1U76
 
 | |
3LZ0
 
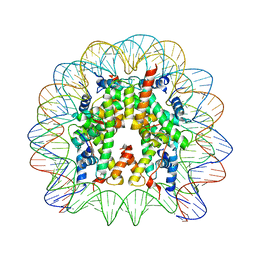 | | Crystal Structure of Nucleosome Core Particle Composed of the Widom 601 DNA Sequence (orientation 1) | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2A, ... | | Authors: | Vasudevan, D, Chua, E.Y.D, Davey, C.A. | | Deposit date: | 2010-03-01 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of nucleosome core particles containing the '601' strong positioning sequence
J.Mol.Biol., 403, 2010
|
|
3ROC
 
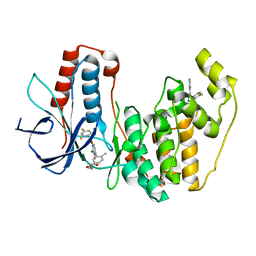 | | Crystal structure of human p38 alpha complexed with a pyrimidinone compound | | Descriptor: | 3-{5-chloro-4-[(2,4-difluorobenzyl)oxy]-6-oxopyrimidin-1(6H)-yl}-N-(2-hydroxyethyl)-4-methylbenzamide, 4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Shieh, H.-S, Xing, L. | | Deposit date: | 2011-04-25 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substituted N-aryl-6-pyrimidinones: A new class of potent, selective, and orally active p38 MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4GIW
 
 | |
3LWI
 
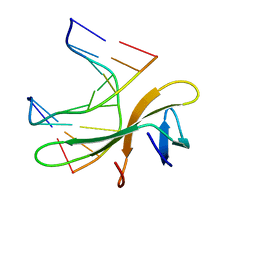 | | Crystal structure of Cren7-dsDNA complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y, Guo, L, Jiang, T, Huang, L. | | Deposit date: | 2010-02-23 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the interaction of the crenarchaeal chromatin protein Cren7 with DNA
Mol.Microbiol., 76, 2010
|
|
3LZ1
 
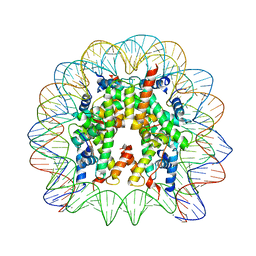 | | Crystal Structure of Nucleosome Core Particle Composed of the Widom 601 DNA Sequence (orientation 2) | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2A, ... | | Authors: | Vasudevan, D, Chua, E.Y.D, Davey, C.A. | | Deposit date: | 2010-03-01 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of nucleosome core particles containing the '601' strong positioning sequence
J.Mol.Biol., 403, 2010
|
|
6TWL
 
 | |
6UNE
 
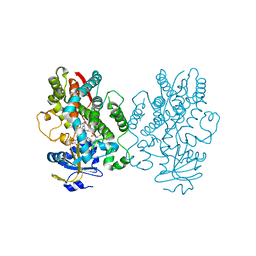 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2R)-1-(1H-indol-3-yl)-3-{[(2S)-3-oxo-2-(phenylamino)-3-{[(pyridin-3-yl)methyl]amino}propyl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2019-10-11 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | An increase in side-group hydrophobicity largely improves the potency of ritonavir-like inhibitors of CYP3A4.
Bioorg.Med.Chem., 28, 2020
|
|
3LWH
 
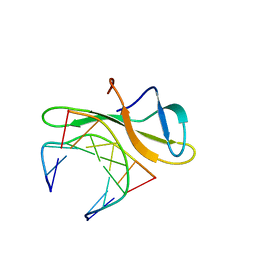 | | Crystal structure of Cren7-dsDNA complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y, Guo, L, Jiang, T, Huang, L. | | Deposit date: | 2010-02-23 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the interaction of the crenarchaeal chromatin protein Cren7 with DNA
Mol.Microbiol., 76, 2010
|
|
2ZVK
 
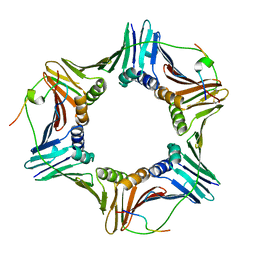 | | Crystal structure of PCNA in complex with DNA polymerase eta fragment | | Descriptor: | DNA polymerase eta, Proliferating cell nuclear antigen | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
6UNK
 
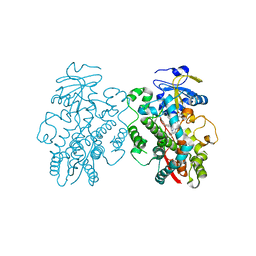 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-(naphthalen-1-yl)-3-{[(2R)-1-oxo-3-phenyl-1-{[2-(pyridin-3-yl)ethyl]amino}propan-2-yl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2019-10-12 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | An increase in side-group hydrophobicity largely improves the potency of ritonavir-like inhibitors of CYP3A4.
Bioorg.Med.Chem., 28, 2020
|
|
1S3O
 
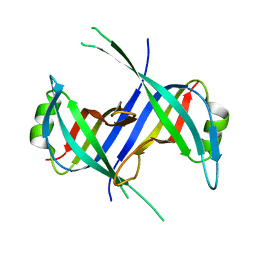 | |
6UNH
 
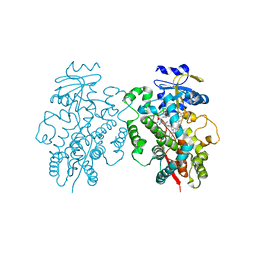 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2R)-1-(1H-indol-3-yl)-3-{[(2S)-1-oxo-3-phenyl-1-{[2-(pyridin-3-yl)ethyl]amino}propan-2-yl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2019-10-11 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | An increase in side-group hydrophobicity largely improves the potency of ritonavir-like inhibitors of CYP3A4.
Bioorg.Med.Chem., 28, 2020
|
|
4HKK
 
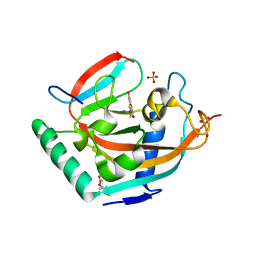 | | Complex structure of human tankyrase 2 with apigenin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2012-10-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Screening and structural analysis of flavones inhibiting tankyrases.
J.Med.Chem., 56, 2013
|
|
4HLK
 
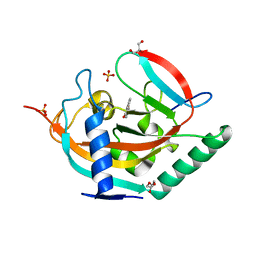 | | Crystal structure of Tankyrase 2 in complex with 4'-methylflavone | | Descriptor: | 2-(4-methylphenyl)-4H-chromen-4-one, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2012-10-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening and structural analysis of flavones inhibiting tankyrases.
J.Med.Chem., 56, 2013
|
|
3NNU
 
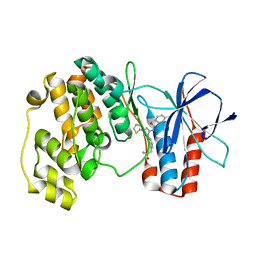 | | Crystal structure of P38 alpha in complex with DP1376 | | Descriptor: | 2-{3-[(5E)-5-{[(2,3-dichlorophenyl)carbamoyl]imino}-3-thiophen-2-yl-2,5-dihydro-1H-pyrazol-1-yl]phenyl}acetamide, Mitogen-activated protein kinase 14 | | Authors: | Abendroth, J. | | Deposit date: | 2010-06-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Switch control pocket inhibitors of p38-MAP kinase. Durable type II inhibitors that do not require binding into the canonical ATP hinge region
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3RIN
 
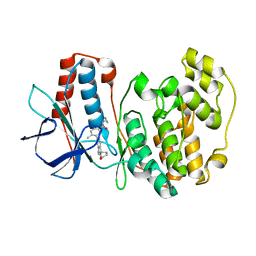 | | p38 kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-4-methyl-3-(2'-oxo-1',2'-dihydrospiro[cyclopentane-1,3'-indol]-6'-yl)benzamide | | Authors: | Segarra, V, Eastwood, P, Roca, R, Fisher, M, Lamers, M. | | Deposit date: | 2011-04-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Indolin-2-one p38(alpha) inhibitors I: design, profiling and crystallographic binding mode.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5EVC
 
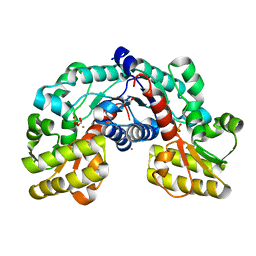 | | Crystal structure of putative aspartate racemase from Salmonella Typhimurium complexed with sulfate and potassium | | Descriptor: | CHLORIDE ION, FLUORIDE ION, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative aspartate racemase from Salmonella Typhimurium complexed with sulfate and potassium
To be published
|
|
3NNW
 
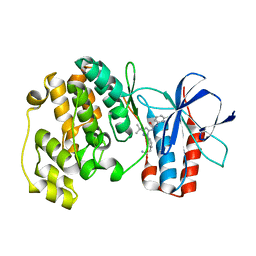 | | Crystal structure of P38 alpha in complex with DP802 | | Descriptor: | 2-[3-(3-tert-butyl-5-{[(2,3-dichlorophenyl)carbamoyl]imino}-2,5-dihydro-1H-pyrazol-1-yl)phenyl]acetamide, Mitogen-activated protein kinase 14 | | Authors: | Abendroth, J. | | Deposit date: | 2010-06-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Switch control pocket inhibitors of p38-MAP kinase. Durable type II inhibitors that do not require binding into the canonical ATP hinge region
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5IBH
 
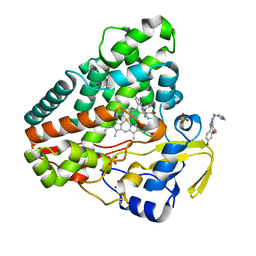 | |
3NNX
 
 | | Crystal structure of phosphorylated P38 alpha in complex with DP802 | | Descriptor: | 2-[3-(3-tert-butyl-5-{[(2,3-dichlorophenyl)carbamoyl]imino}-2,5-dihydro-1H-pyrazol-1-yl)phenyl]acetamide, Mitogen-activated protein kinase 14 | | Authors: | Abendroth, J. | | Deposit date: | 2010-06-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Switch control pocket inhibitors of p38-MAP kinase. Durable type II inhibitors that do not require binding into the canonical ATP hinge region
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5LJM
 
 | | Structure of SPATA2 PUB domain | | Descriptor: | GLYCEROL, Spermatogenesis-associated protein 2 | | Authors: | Elliott, P.R, Komander, D. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling.
Mol.Cell, 63, 2016
|
|
7Z3Y
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH013545 | | Descriptor: | 2-[4-(3,5-dimethylpyrazol-1-yl)-2,6-bis(fluoranyl)phenyl]-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)ethanamide, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Scaletti, E.R, Stenmark, P. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH013545
To Be Published
|
|
7Z5B
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH013546 | | Descriptor: | 2-[4-(2,4-dimethyl-1~{H}-imidazol-5-yl)phenyl]-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)ethanamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Scaletti, E, Stenmark, P. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH013546
To Be Published
|
|
