1OM5
 
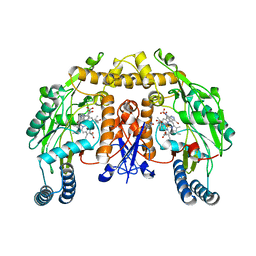 | | STRUCTURE OF RAT NEURONAL NOS HEME DOMAIN WITH 3-BROMO-7-NITROINDAZOLE BOUND | | Descriptor: | 3-BROMO-7-NITROINDAZOLE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L, Raman, C.S. | | Deposit date: | 2003-02-24 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Rat Neuronal NOS Heme Domain with 3-Bromo-7-Nitroindazole Bound
To be Published
|
|
1P6I
 
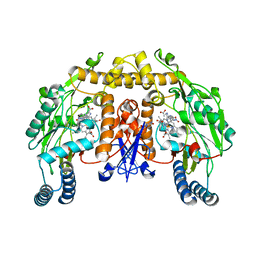 | | Rat neuronal NOS heme domain with (4S)-N-(4-amino-5-[aminoethyl]aminopentyl)-N'-nitroguanidine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(4S)-4-AMINO-5-[(2-AMINOETHYL)AMINO]PENTYL}-N'-NITROGUANIDINE, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1XCV
 
 | |
1E5J
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE TETRAGONAL CRYSTAL FORM IN COMPLEX WITH METHYL-4II-S-ALPHA-CELLOBIOSYL-4II-THIO-BETA-CELLOBIOSIDE | | Descriptor: | CALCIUM ION, ENDOGLUCANASE 5A, alpha-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside | | Authors: | Fort, S, Varrot, A, Schulein, M, Cottaz, S, Driguez, H, Davies, G.J. | | Deposit date: | 2000-07-26 | | Release date: | 2001-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mixed-Linkage Cellooligosaccharides: A New Class of Glycoside Hydrolase Inhibitors
Chembiochem, 2, 2001
|
|
1P6J
 
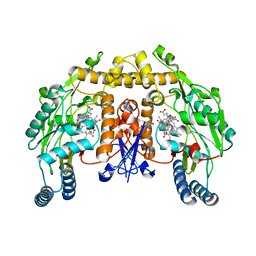 | | Rat neuronal NOS heme domain with L-N(omega)-nitroarginine-(4R)-amino-L-proline amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, L-N(OMEGA)-NITROARGININE-(4R)-AMINO-L-PROLINE AMIDE, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1XC9
 
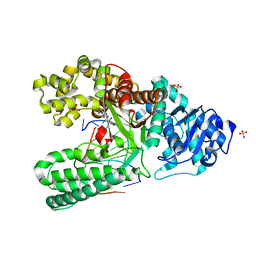 | | Structure of a high-fidelity polymerase bound to a benzo[a]pyrene adduct that blocks replication | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA polymerase I, DNA primer strand, ... | | Authors: | Hsu, G.W, Huang, X, Luneva, N.P, Geacintov, N.E, Beese, L.S. | | Deposit date: | 2004-09-01 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a High Fidelity DNA Polymerase Bound to a Benzo[a]pyrene Adduct That Blocks Replication
J.Biol.Chem., 280, 2005
|
|
1P6M
 
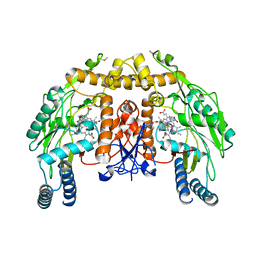 | | Bovine endothelial NOS heme domain with (4S)-N-(4-amino-5-[aminoethyl]aminopentyl)-N'-nitroguanidine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1P84
 
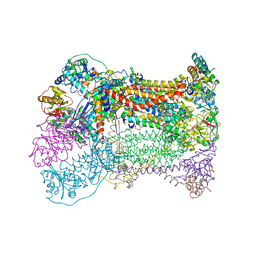 | | HDBT inhibited Yeast Cytochrome bc1 Complex | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Palsdottir, H, Lojero, C.G, Trumpower, B.L, Hunte, C. | | Deposit date: | 2003-05-06 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the yeast cytochrome bc1 complex with a hydroxyquinone anion Qo site inhibitor bound
J.Biol.Chem., 278, 2003
|
|
9C5P
 
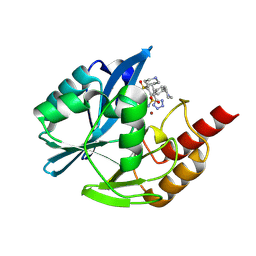 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4-(piperidine-4-sulfonyl)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, MAGNESIUM ION, Metallo-beta-lactamase type 2, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2024-06-06 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Discovery of sulfone containing metallo-beta-lactamase inhibitors with reduced bacterial cell efflux and histamine release issues
BMCL, 2024
|
|
1PK9
 
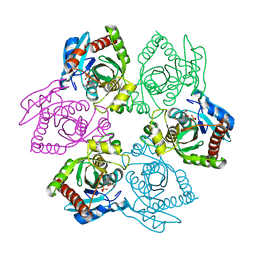 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 2-fluoroadenosine and sulfate/phosphate | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-05 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PKE
 
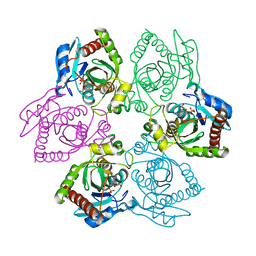 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 2-fluoro-2'-deoxyadenosine and sulfate/phosphate | | Descriptor: | 5-(6-AMINO-2-FLUORO-PURIN-9-YL)-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-OL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-05 | | Release date: | 2003-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
8K1X
 
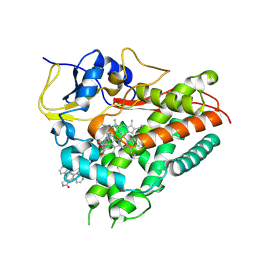 | | Biochemical and structural characterization of a multifunctional cytochrome P450 SpcN in staurosporine biosynthesis | | Descriptor: | 13-(3-Amino-2,3,6-trideoxy-alpha-L-ribo-hexopyranosyl)-6,7,12,13-tetrahydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazol-5-one, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2023-07-11 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular Basis for the P450-Catalyzed sp3 C-N Glycosidic Bond Formation in Staurosporine Biosynthesis.
Acs Catalysis, 14, 2024
|
|
7HIT
 
 | | Group deposition of Chikungunya virus nsP3 macrodomain in complex with inhibitors from the READDI-AC AViDD center -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with RA-0188454-02 (CHIKV_MacB-x2370) | | Descriptor: | 2-methyl-N-{[4-(2H-tetrazol-5-yl)-1,3-thiazol-2-yl]methyl}-1H-pyrrole-3-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Almahli, H, Balcomb, B.H, Capkin, E, Chandran, A.V, Chen, W, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Saleem, R.S.Z, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Todd, M.H, Fearon, D, von Delft, F. | | Deposit date: | 2024-10-04 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Group deposition of Chikungunya virus nsP3 macrodomain in complex with inhibitors from the READDI-AC AViDD center
To Be Published
|
|
7HIB
 
 | | Group deposition of Chikungunya virus nsP3 macrodomain in complex with inhibitors -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with ASAP-0015381-001 (CHIKV_MacB-x2183) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-10-02 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Group deposition of Chikungunya virus nsP3 macrodomain in complex with SARS-CoV-2 NSP3 Macrodomain inhibitors
To Be Published
|
|
1PW7
 
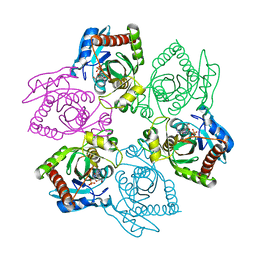 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 9-beta-D-arabinofuranosyladenine and sulfate/phosphate | | Descriptor: | 2-(6-AMINO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-30 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PZK
 
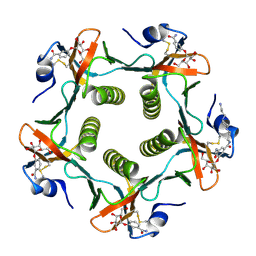 | | Cholera Toxin B-Pentamer Complexed With N-Acyl Phenyl Galactoside 9h | | Descriptor: | Cholera Toxin B Subunit, N-{3-[4-(3-AMINO-PROPYL)-PIPERAZIN-1-YL]-PROPYL}-3-(2-THIOPHEN-2-YL-ACETYLAMINO)-5-(3,4,5-TRIHYDROXY-6-HYDROXYMETHYL-TETRAHYDRO-PYRAN-2-YLOXY)-BENZAMIDE | | Authors: | Mitchell, D.D, Pickens, J.C, Korotkov, K, Fan, E, Hol, W.G.J. | | Deposit date: | 2003-07-11 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 3,5-Substituted phenyl galactosides as leads in designing effective cholera toxin antagonists; synthesis and crystallographic studies
Bioorg.Med.Chem., 12, 2004
|
|
9DTA
 
 | | Crystal structure of the WDR domain of WDR91 in complex with DR3448 | | Descriptor: | N-(1-benzoyl-1,2,3,4-tetrahydroquinolin-6-yl)-2-(3-cyano-2-methoxyphenyl)acetamide, WD repeat-containing protein 91 | | Authors: | Zeng, H, Ahmad, H, Dong, A, Seitova, A, Counago, R.M, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-09-30 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the WDR domain of WDR91 in complex with DR3448
To be published
|
|
1YV3
 
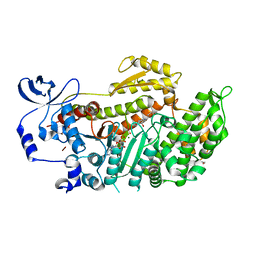 | | The structural basis of blebbistatin inhibition and specificity for myosin II | | Descriptor: | (-)-1-PHENYL-1,2,3,4-TETRAHYDRO-4-HYDROXYPYRROLO[2,3-B]-7-METHYLQUINOLIN-4-ONE, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Allingham, J.S, Smith, R, Rayment, I. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis of blebbistatin inhibition and specificity for myosin II.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1PR4
 
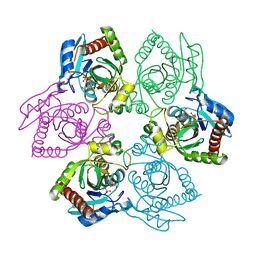 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 9-beta-D-ribofuranosyl-6-methylthiopurine and Phosphate/Sulfate | | Descriptor: | 2-HYDROXYMETHYL-5-(6-METHYLSULFANYL-PURIN-9-YL)-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
9C80
 
 | | Co-structure of SARS-CoV-2 (COVID-19 with covalent inhibitor | | Descriptor: | (5R,7S,8R)-7-(2-fluorophenyl)-3-[(2-fluorophenyl)carbamoyl]-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-5-carboxylic acid, 3C-like proteinase nsp5 | | Authors: | Ornelas, E, Knapp, M.S. | | Deposit date: | 2024-06-11 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Identification of Potent, Broad-Spectrum Coronavirus Main Protease Inhibitors for Pandemic Preparedness.
J.Med.Chem., 2024
|
|
143D
 
 | |
1YLS
 
 | | Crystal structure of selenium-modified Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | Descriptor: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, MAGNESIUM ION, RNA Diels-Alder ribozyme | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YKV
 
 | | Crystal structure of the Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | Descriptor: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, Diels-Alder ribozyme, MAGNESIUM ION | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1Q83
 
 | | Crystal structure of the mouse acetylcholinesterase-TZ2PA6 syn complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,8-DIAMINO-6-PHENYL-5-[6-[1-[2-[(1,2,3,4-TETRAHYDRO-9-ACRIDINYL)AMINO]ETHYL]-1H-1,2,3-TRIAZOL-5-YL]HEXYL]-PHENANTHRIDINIUM, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Kolb, H.C, Radic, Z, Sharpless, K.B, Taylor, P, Marchot, P. | | Deposit date: | 2003-08-20 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Freeze-frame inhibitor captures acetylcholinesterase in a unique conformation.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1O4F
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU79073. | | Descriptor: | 1,2,3,4-TETRAHYDROQUINOLIN-8-YL DIHYDROGEN PHOSPHATE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
