5U5Z
 
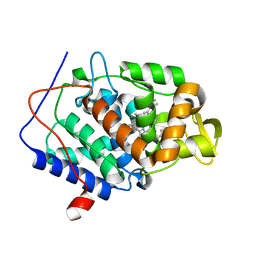 | | CcP gateless cavity | | Descriptor: | 4-methyl-2-phenyl-1H-imidazole, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase | | Authors: | Fischer, M, Shoichet, B.K. | | Deposit date: | 2016-12-07 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Testing inhomogeneous solvation theory in structure-based ligand discovery.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5GTC
 
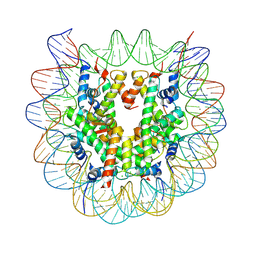 | | Crystal structure of complex between DMAP-SH conjugated with a Kaposi's sarcoma herpesvirus LANA peptide (5-15) and nucleosome core particle | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Kato, D, Suto, H, Kurumizaka, H, Kawashima, S.A, Yamatsugu, K, Kanai, M. | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic Posttranslational Modifications: Chemical Catalyst-Driven Regioselective Histone Acylation of Native Chromatin.
J. Am. Chem. Soc., 139, 2017
|
|
7ULQ
 
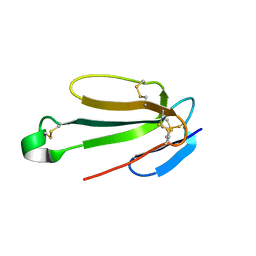 | | Recombinant Hannalgesin | | Descriptor: | ACETATE ION, GLYCEROL, Long neurotoxin OH-55, ... | | Authors: | Xu, J, Lei, X, Chen, L. | | Deposit date: | 2022-04-05 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of recombinant Hannalgesin at 2.2 Angstroms resolution.
To Be Published
|
|
8U8W
 
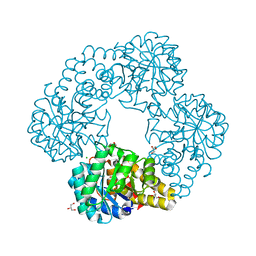 | |
5TZP
 
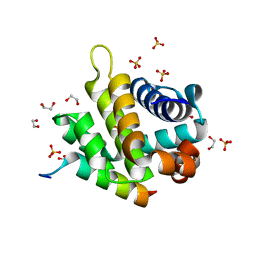 | | Crystal structure of FPV039:Bik BH3 complex | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2-like protein FPV039, Bik, ... | | Authors: | Anasir, M.I, Kvansakul, M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis of apoptosis inhibition by the fowlpox virus protein FPV039.
J. Biol. Chem., 292, 2017
|
|
5G4Y
 
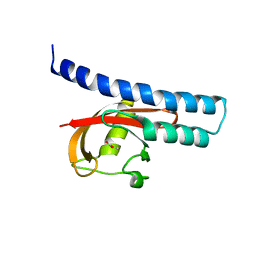 | | Structural basis for carboxylic acid recognition by a Cache chemosensory domain. | | Descriptor: | Methyl-accepting chemotaxis sensory transducer with Cache sensor, UNKNOWN LIGAND | | Authors: | Brewster, J, McKellar, J.L.O, Newman, J, Peat, T.S, Gerth, M.L. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for ligand recognition by a Cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae.
Sci Rep, 6, 2016
|
|
8JS6
 
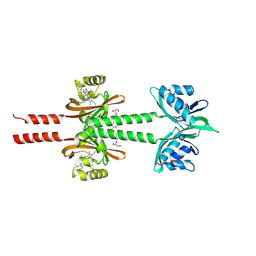 | | Dimeric PAS domains of oxygen sensor FixL in complex with cyanide-bound ferric heme | | Descriptor: | CYANIDE ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kamaya, M, Koteishi, H, Sawai, H, Sugimoto, H, Shiro, Y. | | Deposit date: | 2023-06-19 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dimeric PAS domains of oxygen sensor FixL in complex with imidazole-bound heme.
To be published
|
|
8U97
 
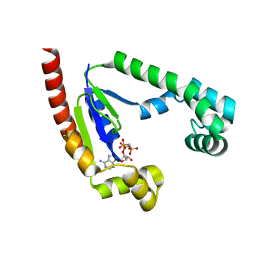 | |
5C6C
 
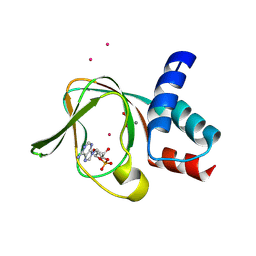 | | PKG II's Amino Terminal Cyclic Nucleotide Binding Domain (CNB-A) in a complex with cAMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CADMIUM ION, ... | | Authors: | Campbell, J.C, Reger, A.S, Huang, G.Y, Sankaran, B, Kim, J.J, Kim, C.W. | | Deposit date: | 2015-06-22 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II.
J.Biol.Chem., 291, 2016
|
|
7P6F
 
 | | 1.93 A resolution X-ray crystal structure of the transcriptional regulator SrnR from Streptomyces griseus | | Descriptor: | ACETATE ION, SODIUM ION, Transcriptional regulator SrnR | | Authors: | Mazzei, L, Ciurli, S. | | Deposit date: | 2021-07-16 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure, dynamics, and function of SrnR, a transcription factor for nickel-dependent gene expression.
Metallomics, 13, 2021
|
|
5G57
 
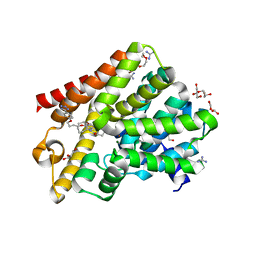 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-001 | | Descriptor: | (4~{a}~{S},8~{a}~{R})-2-cycloheptyl-4-[4-methoxy-3-[4-[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenoxy]butoxy]phenyl]-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2016-05-22 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
5C6K
 
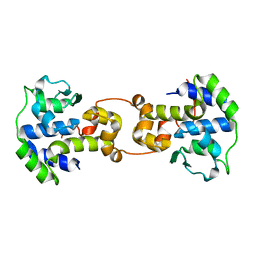 | | Bacteriophage P2 integrase catalytic domain | | Descriptor: | Integrase | | Authors: | Skaar, K, Claesson, M, Odegrip, R, Eriksson, J, Hogbom, M, Haggard-Ljungquist, E, Stenmark, P. | | Deposit date: | 2015-06-23 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the bacteriophage P2 integrase catalytic domain.
Febs Lett., 589, 2015
|
|
7MQT
 
 | |
8QW3
 
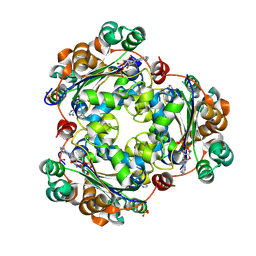 | | Human NDPK-C in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleoside diphosphate kinase 3, SULFATE ION | | Authors: | Werten, S, Amjadi, R, Scheffzek, K. | | Deposit date: | 2023-10-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.255 Å) | | Cite: | Mechanistic Insights into Substrate Recognition of Human Nucleoside Diphosphate Kinase C Based on Nucleotide-Induced Structural Changes.
Int J Mol Sci, 25, 2024
|
|
7UW3
 
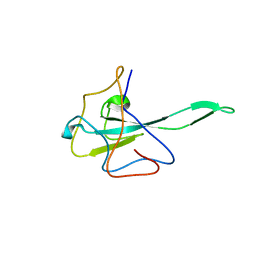 | |
7PAC
 
 | | The crystal structure of PDE6D in the apo state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NICKEL (II) ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Beaumont, E, Williams, D. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Validation of a small molecule inhibitor of PDE6D-RAS interaction with favorable anti-leukemic effects.
Blood Cancer J, 12, 2022
|
|
7UXV
 
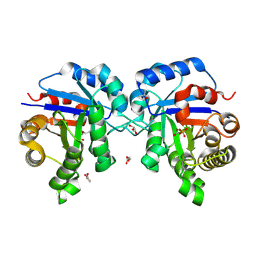 | | human triosephosphate isomerase mutant G122R | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Romero, J.M. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | human triosephosphate isomerase mutant G122R
To Be Published
|
|
8UF5
 
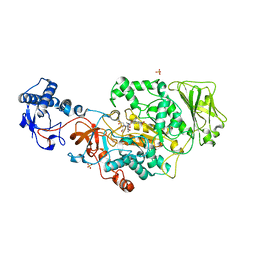 | | Catalytic domain of GtfB in complex with inhibitor G43 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Schormann, N, Deivanayagam, C, Velu, S. | | Deposit date: | 2023-10-03 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Discovery of Small Molecule Inhibitors of Cariogenic Virulence.
Sci Rep, 7, 2017
|
|
7MFN
 
 | |
7OPT
 
 | | Crystal structure of Trypanosoma cruzi peroxidase | | Descriptor: | Ascorbate peroxidase, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Freeman, S.L, Kwon, H, Skafar, V, Fielding, A.J, Martinez, A, Piacenza, L, Radi, R, Raven, E.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of Trypanosoma cruzi heme peroxidase and characterization of its substrate specificity and compound I intermediate.
J.Biol.Chem., 298, 2022
|
|
5TOH
 
 | | Crystal Structure of the Marburg Virus VP35 Oligomerization Domain I2 | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Bruhn, J.F, Kirchdoerfer, R.N, Tickle, I.J, Bricogne, G, Saphire, E.O. | | Deposit date: | 2016-10-17 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of the Marburg Virus VP35 Oligomerization Domain.
J. Virol., 91, 2017
|
|
5GUT
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - N1248A | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Chen, S.J, Ye, F. | | Deposit date: | 2016-08-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
8UC2
 
 | | Ethylene forming enzyme (EFE) R171A variant in complex with nickel and Benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, BENZOIC ACID, ... | | Authors: | Chatterjee, S, Rankin, J.A, Hu, J, Hausinger, R. | | Deposit date: | 2023-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, Spectroscopic, and Computational Insights from Canavanine-Bound and Two Catalytically Compromised Variants of the Ethylene-Forming Enzyme.
Biochemistry, 63, 2024
|
|
7Y0S
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-tyrosyl-L-tyrosine and hydroxylamine | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, I7X-TYR-TYR, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-06-06 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-tyrosyl-L-tyrosine and hydroxylamine
To Be Published
|
|
8QJP
 
 | | SmNuc1 nuclease from Stenotrophomonas maltophilia in complex with uridine - 5'- monophosphate | | Descriptor: | GLYCEROL, PHOSPHATE ION, S1/P1 Nuclease, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Dohnalek, J. | | Deposit date: | 2023-09-13 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate preference, RNA binding and active site versatility of Stenotrophomonas maltophilia nuclease SmNuc1, explained by a structural study.
Febs J., 2024
|
|
