1IDE
 
 | | ISOCITRATE DEHYDROGENASE Y160F MUTANT STEADY-STATE INTERMEDIATE COMPLEX (LAUE DETERMINATION) | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
6XRU
 
 | | GTP-specific succinyl-CoA synthetase complexed with desulfo-coenzyme A, magnesium ions and succinates | | Descriptor: | 1,2-ETHANEDIOL, DESULFO-COENZYME A, GLYCEROL, ... | | Authors: | Huang, J, Fraser, M.E. | | Deposit date: | 2020-07-13 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Second distinct conformation of the phosphohistidine loop in succinyl-CoA synthetase
Acta Crystallogr.,Sect.D, 77, 2021
|
|
8OM7
 
 | | Human Mitochondrial Lon Y186E Mutant ADP Bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
6ILQ
 
 | | Crystal structure of PPARgamma with compound BR101549 | | Descriptor: | Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma, ethyl [2-butyl-6-oxo-1-{[2'-(5-oxo-4,5-dihydro-1,2,4-oxadiazol-3-yl)[1,1'-biphenyl]-4-yl]methyl}-4-(propan-2-yl)-1,6-dihydropyrimidin-5-yl]acetate | | Authors: | Hong, E, Jang, T.H, Chin, J, Kim, K.H, Jung, W, Kim, S.H. | | Deposit date: | 2018-10-19 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.408 Å) | | Cite: | Identification of BR101549 as a lead candidate of non-TZD PPAR gamma agonist for the treatment of type 2 diabetes: Proof-of-concept evaluation and SAR.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3N99
 
 | | Crystal structure of TM1086 | | Descriptor: | CHLORIDE ION, uncharacterized protein TM1086 | | Authors: | Chruszcz, M, Domagalski, M.J, Wang, S, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-28 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of TM1086
To be Published
|
|
5XB6
 
 | |
7Z4W
 
 | | gp6/gp15/gp16 connector complex of bacteriophage SPP1 | | Descriptor: | Head completion protein gp15, Head completion protein gp16, Portal protein | | Authors: | Orlov, I, Roche, S, Tavares, P, Orlova, E.V. | | Deposit date: | 2022-03-05 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | CryoEM structure and assembly mechanism of a bacterial virus genome gatekeeper.
Nat Commun, 13, 2022
|
|
8OUO
 
 | | Human TPC2 in Complex with Antagonist (S)-SG-094 | | Descriptor: | (1S)-6-methoxy-2-methyl-7-phenoxy-1-[(4-phenoxyphenyl)methyl]-3,4-dihydro-1H-isoquinoline, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Chi, G, Pike, A.C.W, Maclean, E.M, Li, H, Mukhopadhyay, S.M.M, Bohstedt, T, Wang, D, McKinley, G, Fernandez-Cid, A, Duerr, K. | | Deposit date: | 2023-04-24 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for inhibition of the lysosomal two-pore channel TPC2 by a small molecule antagonist.
Structure, 2024
|
|
7PFM
 
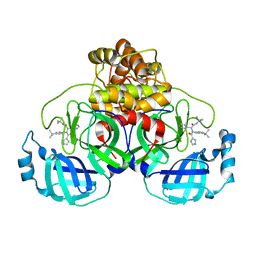 | | A SARS-CoV2 major protease non-covalent ligand structure determined to 2.0 A resolution | | Descriptor: | N-[(1R)-2-(tert-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-N-(4-tert-butylphenyl)-1H-imidazole-5-carboxamide, Replicase polyprotein 1ab | | Authors: | Moche, M, Moodie, L, Strandback, E, Nyman, T, Sandstrom, A, Akaberi, D, Lennerstrand, J. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SARS-CoV2 major protease (Mpro) in complex with a non-covalent inhibitory ligand at 2 A resolution
To Be Published
|
|
3II0
 
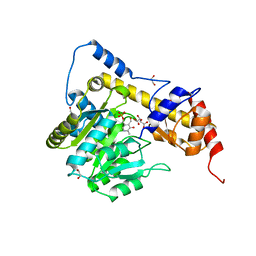 | | Crystal structure of human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | 1,2-ETHANEDIOL, Aspartate aminotransferase, cytoplasmic, ... | | Authors: | Ugochukwu, E, Pilka, E, Cooper, C, Bray, J.E, Yue, W.W, Muniz, J, Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of human Glutamate oxaloacetate transaminase 1 (GOT1)
To be Published
|
|
1F5V
 
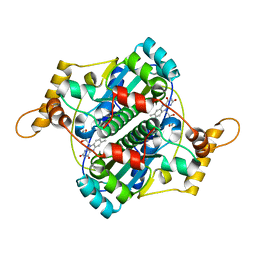 | | STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF A FLAVOPROTEIN FROM ESCHERICHIA COLI THAT REDUCES NITROCOMPOUNDS. ALTERATION OF PYRIDINE NUCLEOTIDE BINDING BY A SINGLE AMINO ACID SUBSTITUTION | | Descriptor: | FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NADPH NITROREDUCTASE | | Authors: | Kobori, T, Sasaki, H, Lee, W.C, Zenno, S, Saigo, K, Murphy, M.E.P, Tanokura, M. | | Deposit date: | 2000-06-17 | | Release date: | 2001-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitrocompounds: alteration of pyridine nucleotide binding by a single amino acid substitution.
J.Biol.Chem., 276, 2001
|
|
1F5W
 
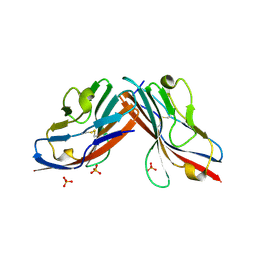 | | DIMERIC STRUCTURE OF THE COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR D1 DOMAIN | | Descriptor: | COXSACKIE VIRUS AND ADENOVIRUS RECEPTOR, SULFATE ION | | Authors: | van Raaij, M.J, Chouin, E, van der Zandt, H, Bergelson, J.M, Cusack, S. | | Deposit date: | 2000-06-18 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 A resolution.
Structure Fold.Des., 8, 2000
|
|
2N87
 
 | |
6AJC
 
 | | Crystal structure of Trypanosoma cruzi cytosolic isocitrate dehydrogenase in complex with NADP+, isocitrate and ca2+ | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP], ... | | Authors: | Wang, X, Inaoka, D.K, Shiba, T, Balogun, E.O, Ziebart, N, Allman, S, Watanabe, Y, Nozaki, T, Boshart, M, Bringaud, F, Harada, S, Kita, K. | | Deposit date: | 2018-08-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical characterization of a novel Trypanosoma brucei glycosomal isocitrate dehydrogenase with dual coenzyme specificity (NADP+/NAD+)
To Be Published
|
|
1FQ4
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN HYDROXYETHYLENE INHIBITOR CP-108,420 AND YEAST ASPARTIC PROTEINASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(2R)-1-{[(2S,3R,5R)-1-cyclohexyl-3-hydroxy-5-{[2-(morpholin-4-yl)ethyl]carbamoyl}oct-7-yn-2-yl]amino}-3-(methylsulfa nyl)-1-oxopropan-2-yl]-1H-benzimidazole-2-carboxamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-03 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
5TMN
 
 | | Slow-and fast-binding inhibitors of thermolysin display different modes of binding. crystallographic analysis of extended phosphonamidate transition-state analogues | | Descriptor: | CALCIUM ION, N-[(S)-({[(benzyloxy)carbonyl]amino}methyl)(hydroxy)phosphoryl]-L-leucyl-L-leucine, THERMOLYSIN, ... | | Authors: | Holden, H.M, Tronrud, D.E, Monzingo, A.F, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Slow- and fast-binding inhibitors of thermolysin display different modes of binding: crystallographic analysis of extended phosphonamidate transition-state analogues.
Biochemistry, 26, 1987
|
|
3IO9
 
 | | BimL12Y in complex with Mcl-1 | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Czabotar, P.E, Lee, E.F, Yang, H, Sleebs, B.E, Lessene, G, Colman, P.M, Smith, B.J, Fairlie, W.D. | | Deposit date: | 2009-08-14 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands.
J.Biol.Chem., 284, 2009
|
|
5IIN
 
 | | Crystal structure of the pre-catalytic ternary extension complex of DNA polymerase lambda with an 8-oxo-dG:dC base-pair | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*(8OG)P*TP*AP*CP*TP*G)-3'), ... | | Authors: | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A fidelity mechanism in DNA polymerase lambda promotes error-free bypass of 8-oxo-dG.
Embo J., 35, 2016
|
|
1F6F
 
 | | CRYSTAL STRUCTURE OF THE TERNARY COMPLEX BETWEEN OVINE PLACENTAL LACTOGEN AND THE EXTRACELLULAR DOMAIN OF THE RAT PROLACTIN RECEPTOR | | Descriptor: | PLACENTAL LACTOGEN, PROLACTIN RECEPTOR | | Authors: | Elkins, P.A, Christinger, H.W, Sandowski, Y, Sakal, E, Gertler, A, De Vos, A.M, Kossiakoff, A.A. | | Deposit date: | 2000-06-21 | | Release date: | 2000-07-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ternary complex between placental lactogen and the extracellular domain of the prolactin receptor.
Nat.Struct.Biol., 7, 2000
|
|
7PKA
 
 | | Synechocystis sp. PCC6803 glutathione transferase Chi 1, GSOH bound | | Descriptor: | Glutathione S-transferase, POTASSIUM ION, S-Hydroxy-Glutathione | | Authors: | Didierjean, C, Mocchetti, E, Hecker, A, Favier, F. | | Deposit date: | 2021-08-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure and functions of glutathione transferase Chi 1 from cyanobacterium Synechocystis sp. PCC 6803
To Be Published
|
|
7PHZ
 
 | | Crystal structure of X77 bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P2(1)2(1)2(1). | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
3NAR
 
 | | Crystal structure of ZHX1 HD4 (zinc-fingers and homeoboxes protein 1, homeodomain 4) | | Descriptor: | SULFATE ION, Zinc fingers and homeoboxes protein 1 | | Authors: | Ren, J, Bird, L.E, Owens, R.J, Stammers, D.K, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel structural features in two ZHX homeodomains derived from a systematic study of single and multiple domains
Bmc Struct.Biol., 10, 2010
|
|
5IIL
 
 | | Crystal structure of the post-catalytic nick complex of DNA polymerase lambda with a templating 8-oxo-dG and incorporated dA | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*AP*GP*TP*AP*AP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*(8OG)P*TP*AP*CP*TP*G)-3'), ... | | Authors: | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A fidelity mechanism in DNA polymerase lambda promotes error-free bypass of 8-oxo-dG.
Embo J., 35, 2016
|
|
6EEB
 
 | | Calmodulin in complex with malbrancheamide | | Descriptor: | (5aS,12aS,13aS)-8,9-dichloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CALCIUM ION, Calmodulin-1, ... | | Authors: | Beyett, T.S, Fraley, A.E, Tesmer, J.J.G. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Perturbation of the interactions of calmodulin with GRK5 using a natural product chemical probe.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4D2T
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 3-[2-(phenylcarbamoyl)-5-(1H-pyrazol-4-yl)phenoxy]propan-1-aminium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-13 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
