4OWW
 
 | | Structural basis of SOSS1 in complex with a 35nt ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), Integrator complex subunit 3, SOSS complex subunit B1, ... | | Authors: | Ren, W, Sun, Q, Tang, X, Song, H. | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of SOSS1 Complex Assembly and Recognition of ssDNA.
Cell Rep, 6, 2014
|
|
7D98
 
 | |
4SKN
 
 | | A NUCLEOTIDE-FLIPPING MECHANISM FROM THE STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*CP*CP*GP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*(D1P)P*GP*GP*CP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE), ... | | Authors: | Slupphaug, G, Mol, C.D, Kavli, B, Arvai, A.S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-02-20 | | Release date: | 1999-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA.
Nature, 384, 1996
|
|
1VKX
 
 | | CRYSTAL STRUCTURE OF THE NFKB P50/P65 HETERODIMER COMPLEXED TO THE IMMUNOGLOBULIN KB DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*C)-3'), PROTEIN (NF-KAPPA B P50 SUBUNIT), ... | | Authors: | Chen, F, Huang, D.B, Ghosh, G. | | Deposit date: | 1997-09-17 | | Release date: | 1998-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA.
Nature, 391, 1998
|
|
5H3R
 
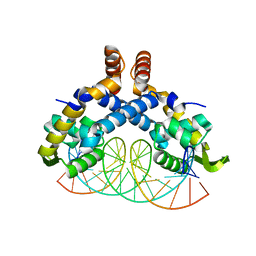 | | Crystal Structure of mutant MarR C80S from E.coli complexed with operator DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*AP*CP*TP*TP*GP*CP*CP*TP*GP*GP*GP*CP*AP*AP*TP*AP*TP*T)-3'), DNA (5'-D(*GP*AP*AP*TP*AP*TP*TP*GP*CP*CP*CP*AP*GP*GP*CP*AP*AP*GP*TP*AP*T)-3'), Multiple antibiotic resistance protein MarR | | Authors: | Zhu, R, Lou, H, Hao, Z. | | Deposit date: | 2016-10-26 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural characterization of the DNA-binding mechanism underlying the copper(II)-sensing MarR transcriptional regulator.
J. Biol. Inorg. Chem., 22, 2017
|
|
1BY4
 
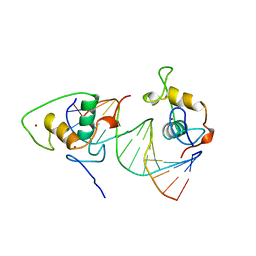 | | STRUCTURE AND MECHANISM OF THE HOMODIMERIC ASSEMBLY OF THE RXR ON DNA | | Descriptor: | DNA (5'-D(*C*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*A)-3'), PROTEIN (RETINOIC ACID RECEPTOR RXR-ALPHA), ... | | Authors: | Zhao, Q, Chasse, S.A, Devarakonda, S, Sierk, M.L, Ahvazi, B, Sigler, P.B, Rastinejad, F. | | Deposit date: | 1998-10-22 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of RXR-DNA interactions.
J.Mol.Biol., 296, 2000
|
|
5FKU
 
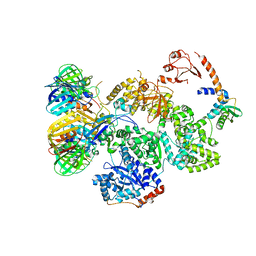 | | cryo-EM structure of the E. coli replicative DNA polymerase complex in DNA free state (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III SUBUNIT ALPHA, DNA POLYMERASE III SUBUNIT BETA, DNA POLYMERASE III SUBUNIT EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.34 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
4LJL
 
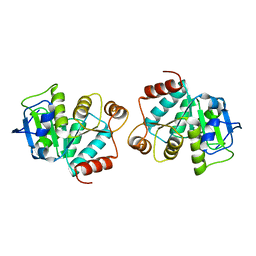 | |
4LJK
 
 | |
3U6Y
 
 | |
7T22
 
 | | E. coli DnaB bound to three DnaG C-terminal domains, ssDNA, ADP and AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA primase, ... | | Authors: | Oakley, A.J, Xu, Z.Q. | | Deposit date: | 2021-12-02 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Sequence of conformation changes of DnaB helicase during DNA unwinding and priming in Escherichia coli
To Be Published
|
|
1EN9
 
 | |
2STW
 
 | | SOLUTION NMR STRUCTURE OF THE HUMAN ETS1/DNA COMPLEX, RESTRAINED REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*TP*CP*GP*AP*AP*CP*TP*TP*CP*CP*GP*GP*CP*TP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*CP*CP*GP*GP*AP*AP*GP*TP*TP*CP*GP*A)-3'), ETS1 | | Authors: | Clore, G.M, Werner, M.H, Gronenborn, A.M. | | Deposit date: | 1996-08-05 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Correction of the NMR structure of the ETS1/DNA complex.
J.Biomol.NMR, 10, 1997
|
|
7PC6
 
 | | DNA-binding domain of a p53 homolog from the hydrothermal vent annelid Alvinella pompejana | | Descriptor: | 1,2-ETHANEDIOL, DNA-binding domain, ZINC ION | | Authors: | Balourdas, D.-I, Knapp, S, Soussi, T, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-08-03 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Evolutionary history of the p53 family DNA-binding domain: insights from an Alvinella pompejana homolog.
Cell Death Dis, 13, 2022
|
|
1EN3
 
 | |
1B72
 
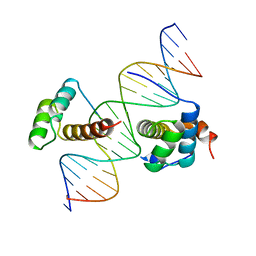 | | PBX1, HOMEOBOX PROTEIN HOX-B1/DNA TERNARY COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*TP*CP*TP*AP*TP*GP*AP*TP*TP*GP*AP*TP*CP*GP*GP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*CP*GP*AP*TP*CP*AP*AP*TP*CP*AP*TP*AP*GP*AP*G)-3'), PROTEIN (HOMEOBOX PROTEIN HOX-B1), ... | | Authors: | Piper, D.E, Batchelor, A.H, Chang, C.-P, Cleary, M.L, Wolberger, C. | | Deposit date: | 1999-01-27 | | Release date: | 1999-02-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation.
Cell(Cambridge,Mass.), 96, 1999
|
|
7VUP
 
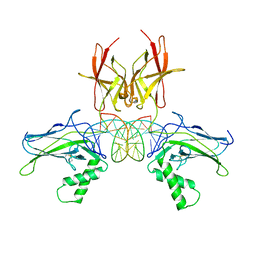 | |
7VUQ
 
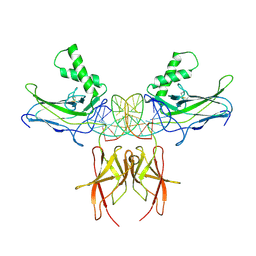 | |
1AZQ
 
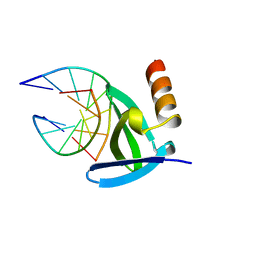 | | HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SAC7D BOUND WITH KINKED DNA DUPLEX | | Descriptor: | DNA (5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3'), PROTEIN (HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SAC7D) | | Authors: | Robinson, H, Gao, Y.-G, Mccrary, B.S, Edmondson, S.P, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1997-11-20 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The hyperthermophile chromosomal protein Sac7d sharply kinks DNA.
Nature, 392, 1998
|
|
1AZP
 
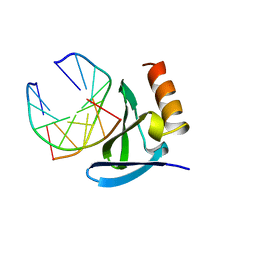 | | HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SAC7D BOUND WITH KINKED DNA DUPLEX | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3'), PROTEIN (HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SAC7D) | | Authors: | Robinson, H, Gao, Y.-G, Mccrary, B.S, Edmondson, S.P, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1997-11-19 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The hyperthermophile chromosomal protein Sac7d sharply kinks DNA.
Nature, 392, 1998
|
|
1YSA
 
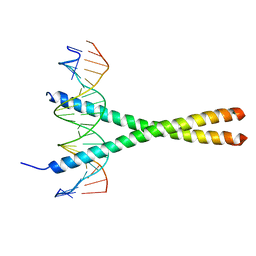 | | THE GCN4 BASIC REGION LEUCINE ZIPPER BINDS DNA AS A DIMER OF UNINTERRUPTED ALPHA HELICES: CRYSTAL STRUCTURE OF THE PROTEIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*CP*TP*GP*GP*AP*TP*GP*AP*GP*TP*CP*AP*TP*A P*GP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*CP*TP*AP*TP*GP*AP*CP*TP*CP*AP*TP*CP*CP*A P*GP*TP*T)-3'), PROTEIN (GCN4) | | Authors: | Ellenberger, T.E, Brandl, C.J, Struhl, K, Harrison, S.C. | | Deposit date: | 1993-08-09 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex.
Cell(Cambridge,Mass.), 71, 1992
|
|
4QJU
 
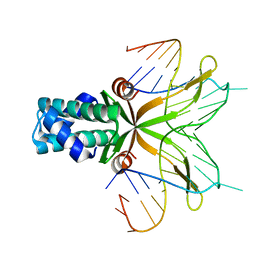 | | Crystal structure of DNA-bound nucleoid associated protein, SAV1473 | | Descriptor: | DNA (5'-D(*TP*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*AP*CP*C)-3'), DNA-binding protein HU | | Authors: | Lee, B.-J, Kim, D.-H, Im, H, Yoon, H.-J. | | Deposit date: | 2014-06-04 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | beta-Arm flexibility of HU from Staphylococcus aureus dictates the DNA-binding and recognition mechanism
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1D87
 
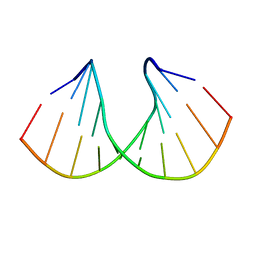 | |
6RKW
 
 | | CryoEM structure of the complete E. coli DNA Gyrase complex bound to a 130 bp DNA duplex | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA (58-MER), DNA (62-MER), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
6RKS
 
 | | E. coli DNA Gyrase - DNA binding and cleavage domain in State 1 without TOPRIM insertion | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA Strand 1, DNA Strand 2, ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
