8E7J
 
 | |
3RY4
 
 | |
5ZGG
 
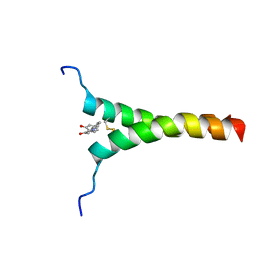 | | NMR structure of p75NTR transmembrane domain in complex with NSC49652 | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-(pyridin-3-yl)prop-2-en-1-one, Tumor necrosis factor receptor superfamily member 16 | | Authors: | Lin, Z, Ibanez, C. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-13 | | Last modified: | 2019-09-25 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Targeting the Transmembrane Domain of Death Receptor p75NTRInduces Melanoma Cell Death and Reduces Tumor Growth.
Cell Chem Biol, 25, 2018
|
|
8AH5
 
 | |
8AH6
 
 | |
8AH8
 
 | |
8AH4
 
 | |
3RY5
 
 | |
6KMP
 
 | | 100K X-ray structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate mimic KVS-1 | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, Protease | | Authors: | Das, A, Kovalevsky, A. | | Deposit date: | 2019-07-31 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
8EFU
 
 | | a22L prion fibril | | Descriptor: | Major prion protein | | Authors: | Hoyt, F, Caughey, B. | | Deposit date: | 2022-09-09 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM of prion strains from the same genotype of host identifies conformational determinants.
Plos Pathog., 18, 2022
|
|
8OVM
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
4HPG
 
 | | Crystal structure of a glycosylated beta-1,3-glucanase (HEV B 2), an allergen from Hevea brasiliensis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-glucanase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rodriguez-Romero, A, Hernandez-Santoyo, A. | | Deposit date: | 2012-10-23 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5364 Å) | | Cite: | Structural analysis of the endogenous glycoallergen Hev b 2 (endo-beta-1,3-glucanase) from Hevea brasiliensis and its recognition by human basophils.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5MGB
 
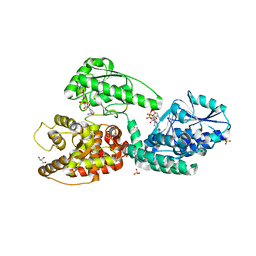 | | Crystal Structure of Rat Peroxisomal Multifunctional enzyme Type-1 (RPMFE1) Complexed with Acetoacetyl-CoA and NAD | | Descriptor: | ACETOACETYL-COENZYME A, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kasaragod, P, Kiema, T.-R, Schmitz, W, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2016-11-21 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural enzymology comparisons of multifunctional enzyme, type-1 (MFE1): the flexibility of its dehydrogenase part.
FEBS Open Bio, 7, 2017
|
|
4QRG
 
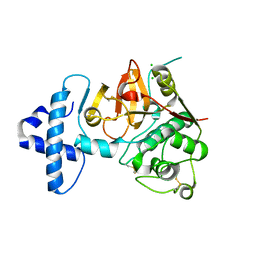 | |
4QRV
 
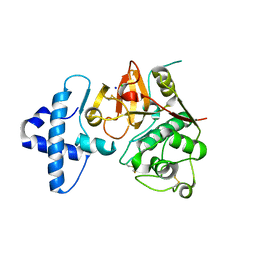 | | Crystal structure of I86F mutant of papain | | Descriptor: | CHLORIDE ION, Papain, SODIUM ION | | Authors: | Dutta, S, Choudhury, D, Roy, S. | | Deposit date: | 2014-07-02 | | Release date: | 2015-08-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Pro-peptide regulates the substrate specificity and zymogen activation process of papain: A structural and mechanistic insight
to be published
|
|
3M7U
 
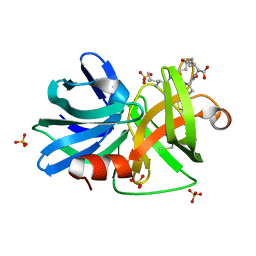 | |
4QRX
 
 | |
4JIX
 
 | | Crystal structure of the metallopeptidase zymogen of Methanocaldococcus jannaschii jannalysin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lopez-Pelegrin, M, Cerda-Costa, N, Martinez-Jimenez, F, Cintas-Pedrola, A, Canals, A, Peinado, J.R, Marti-Renom, M.A, Lopez-Otin, C, Arolas, J.L, Gomis-Ruth, F.X. | | Deposit date: | 2013-03-07 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel family of soluble minimal scaffolds provides structural insight into the catalytic domains of integral membrane metallopeptidases
J.Biol.Chem., 288, 2013
|
|
3M7T
 
 | |
4JIU
 
 | | Crystal structure of the metallopeptidase zymogen of Pyrococcus abyssi abylysin | | Descriptor: | GLYCEROL, Proabylysin, ZINC ION | | Authors: | Lopez-Pelegrin, M, Cerda-Costa, N, Martinez-Jimenez, F, Cintas-Pedrola, A, Canals, A, Peinado, J.R, Marti-Renom, M.A, Lopez-Otin, C, Arolas, J.L, Gomis-Ruth, F.X. | | Deposit date: | 2013-03-07 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A novel family of soluble minimal scaffolds provides structural insight into the catalytic domains of integral membrane metallopeptidases
J.Biol.Chem., 288, 2013
|
|
4TLH
 
 | |
5LRP
 
 | | Mopeia Virus Exonuclease domain complexed with Magnesium | | Descriptor: | MAGNESIUM ION, Nucleoprotein, ZINC ION | | Authors: | Yekwa, E.L, Khourieh, J, Canard, B, Ferron, F. | | Deposit date: | 2016-08-19 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Activity inhibition and crystal polymorphism induced by active-site metal swapping.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5LS4
 
 | | Mopeia virus exonuclease domain complexed with Calcium | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Nucleoprotein, ... | | Authors: | Yekwa, E.L, Canard, B, Ferron, F. | | Deposit date: | 2016-08-22 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.469 Å) | | Cite: | Activity inhibition and crystal polymorphism induced by active-site metal swapping.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5NPT
 
 | | Structure of the N-terminal domain of the yeast telomerase reverse transcriptase | | Descriptor: | Telomerase reverse transcriptase | | Authors: | Rodina, E.V, Lebedev, A.A, Hakanpaa, J, Hackenberg, C, Petrova, O.A, Zvereva, M.I, Dontsova, O.A, Lamzin, V.S. | | Deposit date: | 2017-04-18 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and function of the N-terminal domain of the yeast telomerase reverse transcriptase.
Nucleic Acids Res., 46, 2018
|
|
3NR2
 
 | | Crystal structure of Caspase-6 zymogen | | Descriptor: | Caspase-6 | | Authors: | Su, X.-D, Wang, X.-J, Liu, X, Mi, W, Wang, K.-T. | | Deposit date: | 2010-06-30 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation
Embo Rep., 11, 2010
|
|
