1LHU
 
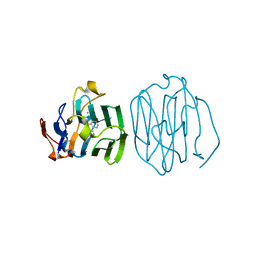 | | CRYSTAL STRUCTURE OF THE N-TERMINAL LG-DOMAIN OF SHBG IN COMPLEX WITH ESTRADIOL | | Descriptor: | CALCIUM ION, ESTRADIOL, SEX HORMONE-BINDING GLOBULIN | | Authors: | Grishkovskaya, I, Avvakumov, G.V, Hammond, G.L, Catalano, M.G, Muller, Y.A. | | Deposit date: | 2002-04-17 | | Release date: | 2002-10-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Steroid Ligands Bind Human Sex Hormone-binding Globulin in Specific Orientations and Produce Distinct Changes in Protein Conformation
J.Biol.Chem., 277, 2002
|
|
2CRA
 
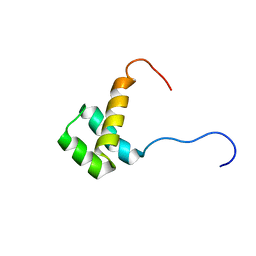 | | Solution structure of the homeobox domain of human homeo box B13 | | Descriptor: | Homeobox protein Hox-B13 | | Authors: | Seimiya, K, Hayashi, F, Nagashima, T, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homeobox domain of human homeo box B13
To be published
|
|
1L7F
 
 | | Crystal structure of influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
2CRM
 
 | |
1LHT
 
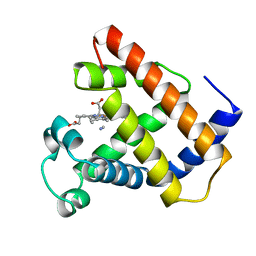 | | LOGGERHEAD SEA TURTLE MYOGLOBIN (CYANO-MET) | | Descriptor: | CYANIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nardini, M, Tarricone, C, Lania, A, Desideri, A, De Sanctis, G, Coletta, M, Petruzzelli, R, Ascenzi, P, Coda, A, Bolognesi, M. | | Deposit date: | 1995-02-01 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reptile heme protein structure: X-ray crystallographic study of the aquo-met and cyano-met derivatives of the loggerhead sea turtle (Caretta caretta) myoglobin at 2.0 A resolution.
J.Mol.Biol., 247, 1995
|
|
1LJE
 
 | |
2WMX
 
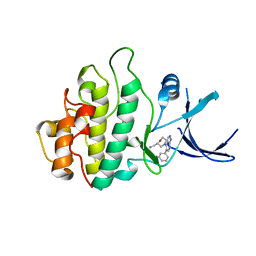 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1-[(2S)-4-(5-phenyl-1H-pyrazolo[3,4-b]pyridin-4-yl)morpholin-2-yl]methanamine, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
2CUM
 
 | | The solution structure of the 33rd fibronectin type III domain of human Tenascin-X | | Descriptor: | Tenascin-X | | Authors: | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-26 | | Release date: | 2005-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the 33rd fibronectin type III domain of human Tenascin-X
To be Published
|
|
2WMQ
 
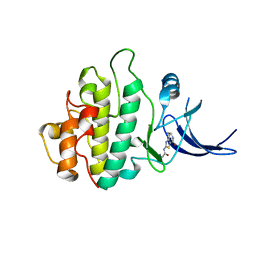 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | N-(4-OXO-5,6,7,8-TETRAHYDRO-4H-[1,3]THIAZOLO[5,4-C]AZEPIN-2-YL)ACETAMIDE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
2CBR
 
 | | CELLULAR RETINOIC ACID BINDING PROTEIN I IN COMPLEX WITH A RETINOBENZOIC ACID (AM80) | | Descriptor: | 4-[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbamoyl]benzoic acid, PROTEIN (CRABP-I) | | Authors: | Chaudhuri, B, Kleywegt, G.J, Bergfors, T, Jones, T.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of cellular retinoic acid binding proteins I and II in complex with synthetic retinoids.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2C8O
 
 | | lysozyme (1sec) and UV lasr excited fluorescence | | Descriptor: | LYSOZYME C | | Authors: | Vernede, X, Lavault, B, Ohana, J, Nurizzo, D, Joly, J, Jacquamet, L, Felisaz, F, Cipriani, F, Bourgeois, D. | | Deposit date: | 2005-12-06 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Uv Laser-Excited Fluorescence as a Tool for the Visualization of Protein Crystals Mounted in Loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1LPU
 
 | | Low Temperature Crystal Structure of the Apo-form of the catalytic subunit of protein kinase CK2 from Zea mays | | Descriptor: | BENZAMIDINE, Protein kinase CK2 | | Authors: | Niefind, K, Puetter, M, Guerra, B, Issinger, O.-G, Schomburg, D. | | Deposit date: | 2002-05-08 | | Release date: | 2002-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Inclining the purine base binding plane in protein kinase CK2 by exchanging the flanking side-chains generates a preference for ATP as a cosubstrate.
J.Mol.Biol., 347, 2005
|
|
1LC1
 
 | |
1TRN
 
 | |
2C8R
 
 | | insuline(60sec) and UV laser excited fluorescence | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Vernede, X, Lavault, B, Ohana, J, Nurizzo, D, Joly, J, Jacquamet, L, Felisaz, F, Cipriani, F, Bourgeois, D. | | Deposit date: | 2005-12-06 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Uv Laser-Excited Fluorescence as a Tool for the Visualization of Protein Crystals Mounted in Loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1LE6
 
 | | CARBOXYLIC ESTER HYDROLASE, P 1 21 1 SPACE GROUP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Group X Secretory Phospholipase A2 | | Authors: | Pan, Y.H, Jain, M.K, Bahnson, B.J. | | Deposit date: | 2002-04-09 | | Release date: | 2002-08-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of human group X secreted phospholipase A2. Electrostatically neutral interfacial surface targets zwitterionic membranes.
J.Biol.Chem., 277, 2002
|
|
1KYN
 
 | | Cathepsin-G | | Descriptor: | (2-NAPHTHALEN-2-YL-1-NAPHTHALEN-1-YL-2-OXO-ETHYL)-PHOSPHONIC ACID, cathepsin G | | Authors: | Greco, M.N, Hawkins, M.J, Powell, E.T, Almond Jr, H.R, Corcoran, T.W, De Garavilla, L, Kauffman, J.A, Recacha, R, Chattopadhyay, D, Andrade-Gordon, P, Maryanoff, B.E. | | Deposit date: | 2002-02-05 | | Release date: | 2002-05-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nonpeptide inhibitors of cathepsin G: optimization of a novel beta-ketophosphonic acid lead by structure-based drug design.
J.Am.Chem.Soc., 124, 2002
|
|
1ZL7
 
 | | Crystal structure of catalytically-active phospholipase A2 with bound calcium | | Descriptor: | CALCIUM ION, GLYCEROL, hypotensive phospholipase A2 | | Authors: | Murakami, M.T, Cintra, A.C, Gabdoulkhakov, A, Genov, N, Betzel, C, Arni, R.K. | | Deposit date: | 2005-05-05 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into metal ion binding in phospholipases A(2): ultra high-resolution crystal structures of an acidic phospholipase A(2) in the Ca(2+) free and bound states.
Biochimie, 88, 2006
|
|
1ZLR
 
 | | Factor XI catalytic domain complexed with 2-guanidino-1-(4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)ethyl nicotinate | | Descriptor: | (1R)-2-{[AMINO(IMINO)METHYL]AMINO}-1-{4-[(4R)-4-(HYDROXYMETHYL)-1,3,2-DIOXABOROLAN-2-YL]PHENYL}ETHYL NICOTINATE, Coagulation factor XI, GLYCEROL, ... | | Authors: | Lazarova, T.I, Jin, L, Rynkiewicz, M, Gorga, J.C, Bibbins, F, Meyers, H.V, Babine, R, Strickler, J. | | Deposit date: | 2005-05-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthesis and in vitro biological evaluation of aryl boronic acids as potential inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1ZM6
 
 | | Crystal structure of the complex formed beween a group I phospholipase A2 and designed penta peptide Leu-Ala-Ile-Tyr-Ser at 2.6A resolution | | Descriptor: | ACETATE ION, Phospholipase A2 isoform 3, designed penta peptide Leu-Ala-Ile-Tyr-Ser | | Authors: | Singh, R.K, Singh, N, Jabeen, T, Sharma, S, Dey, S, Singh, T.P. | | Deposit date: | 2005-05-10 | | Release date: | 2005-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the complex of group I PLA2 with a group II-specific peptide Leu-Ala-Ile-Tyr-Ser (LAIYS) at 2.6 A resolution.
J.Drug Target., 13, 2005
|
|
1LF5
 
 | |
1YVT
 
 | | The high salt (phosphate) crystal structure of CO Hemoglobin E (Glu26Lys) at physiological pH (pH 7.35) | | Descriptor: | CARBON MONOXIDE, GLYCEROL, Hemoglobin alpha chain, ... | | Authors: | Malashkevich, V.N, Balazs, T.C, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2005-02-16 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The high salt (phosphate) crystal structure of CO Hemoglobin E (Glu26Lys) at physiological pH (pH 7.35)
To be Published
|
|
2ZMD
 
 | | Crystal structure of human Mps1 catalytic domain T686A mutant in complex with SP600125 inhibitor | | Descriptor: | 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK | | Authors: | Chu, M.L.H, Chavas, L.M.G, Douglas, K.T, Eyers, P.A, Tabernero, L. | | Deposit date: | 2008-04-16 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of the catalytic domain of the mitotic checkpoint kinase Mps1 in complex with SP600125.
J.Biol.Chem., 283, 2008
|
|
1LGF
 
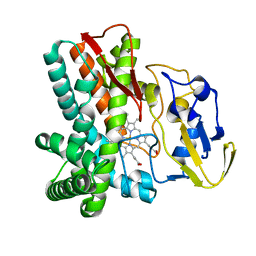 | | Crystal structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction During Vancomycin Biosynthesis | | Descriptor: | P450 monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pylypenko, O, Zerbe, K, Vitali, F, Zhang, W, Vrijbloed, J.W, Robinson, J.A, Schlichting, I. | | Deposit date: | 2002-04-15 | | Release date: | 2002-12-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction during Vancomycin Biosynthesis.
J.Biol.Chem., 277, 2002
|
|
2ZOQ
 
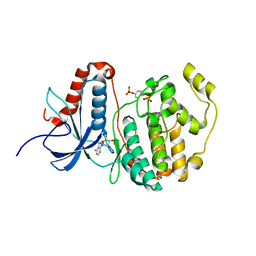 | | Structural dissection of human mitogen-activated kinase ERK1 | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Mitogen-activated protein kinase 3, SODIUM ION, ... | | Authors: | Kinoshita, T, Tada, T, Nakae, S, Yoshida, I. | | Deposit date: | 2008-06-01 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of human mono-phosphorylated ERK1 at Tyr204
Biochem.Biophys.Res.Commun., 377, 2008
|
|
