3MAG
 
 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M3ADE AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 6-AMINO-3-METHYLPURINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1998-07-12 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3MCT
 
 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M3CYT AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 3-METHYLCYTOSINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Gershon, P.D, Hodel, A.E, Quiocho, F.A. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3M0X
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329L in complex with D-psicose | | Descriptor: | D-psicose, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
1E5T
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, MUTANT | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Fulop, V. | | Deposit date: | 2000-08-02 | | Release date: | 2000-10-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalysis of Serine Oligopeptidases is Controlled by a Gating Filter Mechanism
Embo Rep., 1, 2000
|
|
1EAE
 
 | |
1E6L
 
 | | Two-component signal transduction system D13A mutant of CheY | | Descriptor: | Chemotaxis protein CheY | | Authors: | Sola, M, Lopez-Hernandez, E, Cronet, P, Lacroix, E, Serrano, L, Coll, M, Parraga, A. | | Deposit date: | 2000-08-18 | | Release date: | 2001-03-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY.
J.Mol.Biol., 303, 2000
|
|
1E8S
 
 | | Alu domain of the mammalian SRP (potential Alu retroposition intermediate) | | Descriptor: | 7SL RNA, 88-MER, EUROPIUM (III) ION, ... | | Authors: | Weichenrieder, O, Wild, K, Strub, K, Cusack, S. | | Deposit date: | 2000-09-29 | | Release date: | 2000-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure and Assembly of the Alu Domain of the Mammalian Signal Recognition Particle
Nature, 408, 2000
|
|
1E97
 
 | |
1EA2
 
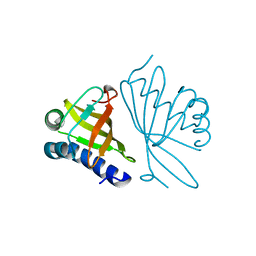 | | Pseudoreversion of the Catalytic Activity of Y14F by the Additional Tyrosine-to-Phenylalanine Substitution(s) in the Hydrogen Bond Network of Delta-5-3-Ketosteroid Isomerase from Pheudomonas putida Biotype B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Choi, G, Ha, N.-C, Kim, M.-S, Hong, B.-H, Choi, K.Y. | | Deposit date: | 2000-11-03 | | Release date: | 2001-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudoreversion of the Catalytic Activity of Y14F by the Additional Substitution(S) of Tyrosine with Phenylalanine in the Hydrogen Bond Network of Delta (5)-3-Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochemistry, 40, 2001
|
|
4UBD
 
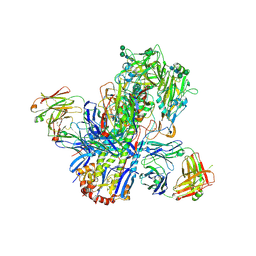 | | Crystal structure of a neutralizing human monoclonal antibody with 1968 H3 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Shore, D.A, Yang, H, Cho, M, Donis, R.O, Stevens, J. | | Deposit date: | 2014-08-12 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus.
Nat Commun, 6, 2015
|
|
1EV8
 
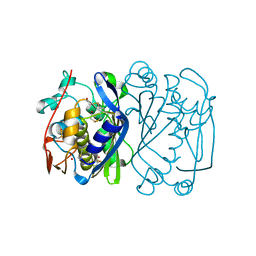 | | CRYSTAL STRUCTURE ANALYSIS OF CYS167 MUTANT OF ESCHERICHIA COLI | | Descriptor: | THYMIDYLATE SYNTHASE | | Authors: | Phan, J, Mahdavian, E, Nivens, M.C, Minor, W, Berger, S, Spencer, H.T, Dunlap, R.B, Lebioda, L. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalytic cysteine of thymidylate synthase is activated upon substrate binding.
Biochemistry, 39, 2000
|
|
1E7U
 
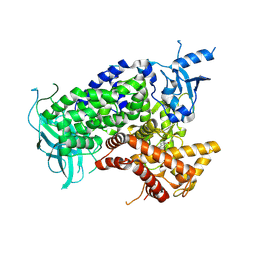 | | Structure determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT | | Authors: | Walker, E.H, Perisic, O, Ried, C, Stephens, L, Williams, R.L. | | Deposit date: | 2000-09-08 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Determinations of Phosphoinositide 3-Kinase Inhibition by Wortmannin, Ly294002, Quercetin, Myricetin and Staurosporine
Mol.Cell, 6, 2000
|
|
1ERU
 
 | | HUMAN THIOREDOXIN (OXIDIZED FORM) | | Descriptor: | THIOREDOXIN | | Authors: | Weichsel, A, Gasdaska, J.R, Powis, G, Montfort, W.R. | | Deposit date: | 1996-02-07 | | Release date: | 1996-08-01 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of reduced, oxidized, and mutated human thioredoxins: evidence for a regulatory homodimer.
Structure, 4, 1996
|
|
1E55
 
 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZMGluE191D in complex with the competitive inhibitor dhurrin | | Descriptor: | (2S)-HYDROXY(4-HYDROXYPHENYL)ETHANENITRILE, BETA-GLUCOSIDASE, beta-D-glucopyranose | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Zamboni, V, Henrissat, B, Esen, A. | | Deposit date: | 2000-07-18 | | Release date: | 2000-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of substrate (aglycone) specificity in beta-glucosidases is revealed by crystal structures of mutant maize beta-glucosidase-DIMBOA, -DIMBOAGlc, and -dhurrin complexes.
Proc. Natl. Acad. Sci. U.S.A., 97, 2000
|
|
1E63
 
 | | Ferredoxin:NADP+ Reductase Mutant with LYS 75 Replaced by SER (K75S) | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Gomez-Moreno, C. | | Deposit date: | 2000-08-07 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Interactions for Complex Formation between Ferredoxin-Nadp+ Reductase and its Protein Partners.
Proteins, 59, 2005
|
|
1ETJ
 
 | | AZURIN MUTANT WITH MET 121 REPLACED BY GLU | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Karlsson, B.G, Tsai, L.-C, Nar, H, Sanders-Loehr, J, Bonander, N, Langer, V, Sjolin, L. | | Deposit date: | 1997-01-11 | | Release date: | 1997-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure determination and characterization of the Pseudomonas aeruginosa azurin mutant Met121Glu.
Biochemistry, 36, 1997
|
|
1E8U
 
 | | Structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HEMAGGLUTININ-NEURAMINIDASE, ... | | Authors: | Crennell, S, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2000-10-01 | | Release date: | 2001-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Multifunctional Paramyxovirus Hemagglutinin-Neuraminidase
Nat.Struct.Biol., 7, 2000
|
|
1EAX
 
 | | Crystal structure of MTSP1 (matriptase) | | Descriptor: | BENZAMIDINE, SULFATE ION, SUPPRESSOR OF TUMORIGENICITY 14 | | Authors: | Friedrich, R, Bode, W. | | Deposit date: | 2001-07-17 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Catalytic Domain Structures of Mt-Sp1/Matriptase, a Matrix-Degrading Transmembrane Serine Proteinase.
J.Biol.Chem., 277, 2002
|
|
1EUS
 
 | | SIALIDASE COMPLEXED WITH 2-DEOXY-2,3-DEHYDRO-N-ACETYLNEURAMINIC ACID | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, SIALIDASE | | Authors: | Gaskell, A, Crennell, S.J, Taylor, G.L. | | Deposit date: | 1996-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll.
Structure, 3, 1995
|
|
1ECJ
 
 | |
1EDM
 
 | | EPIDERMAL GROWTH FACTOR-LIKE DOMAIN FROM HUMAN FACTOR IX | | Descriptor: | CALCIUM ION, FACTOR IX | | Authors: | Rao, Z, Handford, P, Mayhew, M, Knott, V, Brownlee, G.G, Stuart, D. | | Deposit date: | 1996-03-21 | | Release date: | 1996-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of a Ca(2+)-binding epidermal growth factor-like domain: its role in protein-protein interactions.
Cell(Cambridge,Mass.), 82, 1995
|
|
1EI4
 
 | | B-DNA DODECAMER CGCGAAT(TLC)CGCG WITH INCORPORATED [3.3.0]BICYCLO-ARABINO-THYMINE-5'-PHOSPHATE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(TLC)P*(TLC)P*CP*GP*CP*G)-3'), MAGNESIUM ION, SPERMINE | | Authors: | Minasov, G, Teplova, M, Nielsen, P, Wengel, J, Egli, M. | | Deposit date: | 2000-02-24 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural basis of cleavage by RNase H of hybrids of arabinonucleic acids and RNA.
Biochemistry, 39, 2000
|
|
1EYM
 
 | | FK506 BINDING PROTEIN MUTANT, HOMODIMERIC COMPLEX | | Descriptor: | FK506 BINDING PROTEIN | | Authors: | Rollins, C.T, Rivera, V.M, Woolfson, D.N, Keenan, T, Hatada, M, Adams, S.E, Andrade, L.J, Yaeger, D, van Schravendijk, M.R, Holt, D.A, Gilman, M, Clackson, T. | | Deposit date: | 2000-05-07 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A ligand-reversible dimerization system for controlling protein-protein interactions.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1ENA
 
 | |
1EMC
 
 | |
