1JLW
 
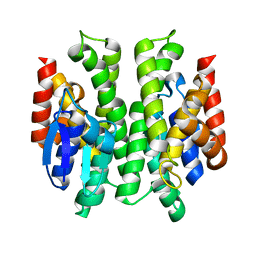 | | Anopheles dirus species B glutathione S-transferases 1-4 | | Descriptor: | glutathione transferase GST1-4 | | Authors: | Oakley, A.J, Harnnoi, T, Udomsinprasert, R, Jirajaroenrat, K, Ketterman, A.J, Wilce, M.C. | | Deposit date: | 2001-07-16 | | Release date: | 2002-07-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The crystal structures of glutathione S-transferases isozymes 1-3 and 1-4 from Anopheles dirus species B.
Protein Sci., 10, 2001
|
|
1JM4
 
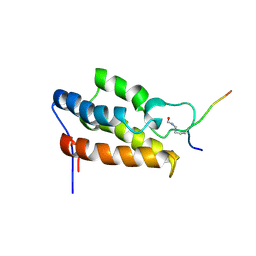 | | NMR Structure of P/CAF Bromodomain in Complex with HIV-1 Tat Peptide | | Descriptor: | HIV-1 Tat Peptide, P300/CBP-associated Factor | | Authors: | Mujtaba, S, He, Y, Zeng, L, Farooq, A, Carlson, J.E, Ott, M, Verdin, E, Zhou, M.-M. | | Deposit date: | 2001-07-17 | | Release date: | 2002-07-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain
Mol.Cell, 9, 2002
|
|
3NTL
 
 | |
5TB6
 
 | | Structure of bromodomain of CREBBP with a pyrazolo[4,3-c]pyridin fragment | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-phenyl-1,4,6,7-tetrahydropyrazolo[4,3-c]pyridin-5-yl)propan-1-one, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Knapp, S, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
9EVZ
 
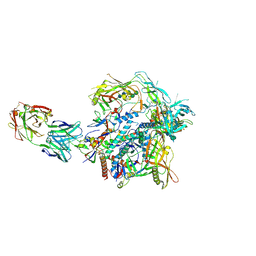 | | HIV-1 envelope glycoprotein (BG505 gp140 SOSIP.664) trimer in complex with ELC07 broadly neutralizing antibody. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ELC07 heavy chain, ... | | Authors: | Hope, J, Alguel, Y, Nans, A, Cherepanov, P. | | Deposit date: | 2024-04-02 | | Release date: | 2025-04-16 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | TITLE: Preservation of memory B cell homeostasis in an individual producing broadly neutralizing antibodies against HIV-1
To Be Published
|
|
1K0N
 
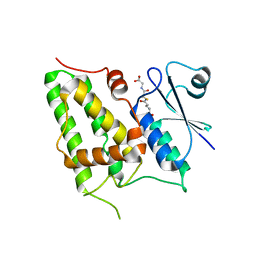 | | Chloride Intracellular Channel 1 (CLIC1) complexed with glutathione | | Descriptor: | CHLORIDE INTRACELLULAR CHANNEL PROTEIN 1, GLUTATHIONE | | Authors: | Harrop, S.J, DeMaere, M.Z, Fairlie, W.D, Reztsova, T, Valenzuela, S.M, Mazzanti, M, Tonini, R, Qiu, M.R, Jankova, L, Warton, K, Bauskin, A.R, Wu, W.M, Pankhurst, S, Campbell, T.J, Breit, S.N, Curmi, P.M.G. | | Deposit date: | 2001-09-19 | | Release date: | 2001-12-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-A resolution.
J.Biol.Chem., 276, 2001
|
|
1K1U
 
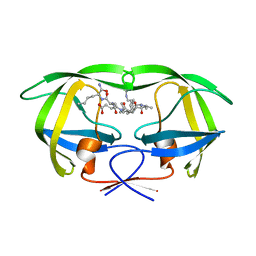 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | Authors: | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, W Harrison, R, Weber, I.T. | | Deposit date: | 2001-09-25 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
5DSY
 
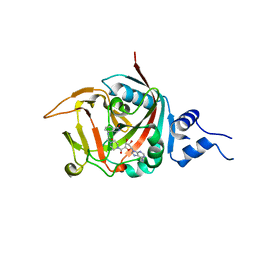 | | Crystal structure of constitutively active PARP-2 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Poly [ADP-ribose] polymerase 2 | | Authors: | Riccio, A.A, Pascal, J.M. | | Deposit date: | 2015-09-17 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain.
Mol.Cell, 60, 2015
|
|
4PY2
 
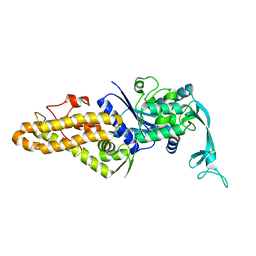 | |
5THJ
 
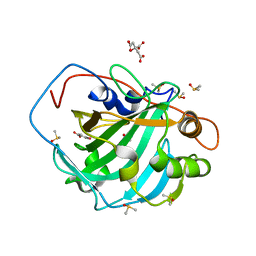 | | Crystal Structure of 2-hydroxycyclohepta-2,4,6-trien-1-one bound to human carbonic anhydrase 2 | | Descriptor: | 2-HYDROXYCYCLOHEPTA-2,4,6-TRIEN-1-ONE, CITRIC ACID, Carbonic anhydrase 2, ... | | Authors: | Dick, B, Cohen, S. | | Deposit date: | 2016-09-29 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effect of donor atom identity on metal-binding pharmacophore coordination.
J. Biol. Inorg. Chem., 22, 2017
|
|
9F02
 
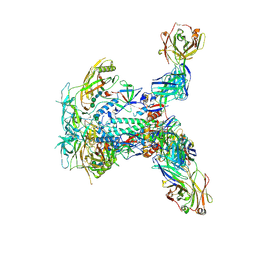 | | HIV-1 envelope glycoprotein (BG505 gp140 SOSIP.664) trimer in complex with three copies of ELC07 broadly neutralizing antibody. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ELC07 heavy chain, ... | | Authors: | Hope, J, Alguel, Y, Nans, A, Cherepanov, P. | | Deposit date: | 2024-04-15 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | TITLE: Preservation of memory B cell homeostasis in an individual producing broadly neutralizing antibodies against HIV-1
To Be Published
|
|
1JTD
 
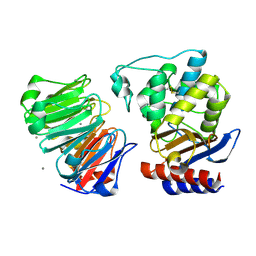 | | Crystal structure of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase | | Descriptor: | CALCIUM ION, TEM-1 beta-lactamase, beta-lactamase inhibitor protein II | | Authors: | Lim, D.C, Park, H.U, De Castro, L, Kang, S.G, Lee, H.S, Jensen, S, Lee, K.J, Strynadka, N.C.J. | | Deposit date: | 2001-08-20 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and kinetic analysis of beta-lactamase inhibitor protein-II in complex with TEM-1 beta-lactamase.
Nat.Struct.Biol., 8, 2001
|
|
1P6O
 
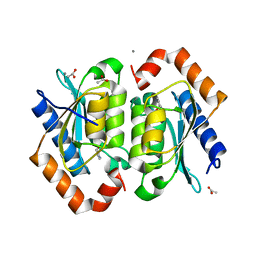 | | The crystal structure of yeast cytosine deaminase bound to 4(R)-hydroxyl-3,4-dihydropyrimidine at 1.14 angstroms. | | Descriptor: | 4-HYDROXY-3,4-DIHYDRO-1H-PYRIMIDIN-2-ONE, ACETIC ACID, CALCIUM ION, ... | | Authors: | Ireton, G.C, Black, M.E, Stoddard, B.L. | | Deposit date: | 2003-04-29 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | The 1.14 a crystal structure of yeast Cytosine deaminase. Evolution of nucleotide salvage enzymes and implications for genetic chemotherapy.
Structure, 11, 2003
|
|
4MNG
 
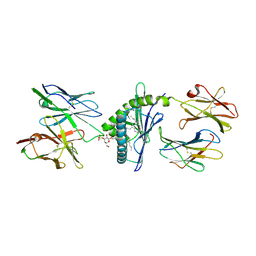 | | Structure of the DP10.7 TCR with CD1d-sulfatide | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Luoma, A.M, Adams, E.J. | | Deposit date: | 2013-09-10 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.0058 Å) | | Cite: | Crystal Structure of V delta 1 T Cell Receptor in Complex with CD1d-Sulfatide Shows MHC-like Recognition of a Self-Lipid by Human gamma delta T Cells.
Immunity, 39, 2013
|
|
5THI
 
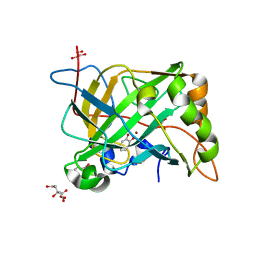 | | Crystal Structure of 2-hydroxycyclohepta-2,4,6-trien-1-one bound to human carbonic anhydrase 2 L198G | | Descriptor: | 2-HYDROXYCYCLOHEPTA-2,4,6-TRIEN-1-ONE, CITRIC ACID, Carbonic anhydrase 2, ... | | Authors: | Dick, B, Cohen, S. | | Deposit date: | 2016-09-29 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effect of donor atom identity on metal-binding pharmacophore coordination.
J. Biol. Inorg. Chem., 22, 2017
|
|
5TI0
 
 | | Crystal Structure of 2-Hydroxycyclohepta-2,4,6-triene-1-thione bound to human carbonic anhydrase 2 L198G | | Descriptor: | 2-hydroxycyclohepta-2,4,6-triene-1-thione, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Dick, B, Cohen, S. | | Deposit date: | 2016-09-30 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Effect of donor atom identity on metal-binding pharmacophore coordination.
J. Biol. Inorg. Chem., 22, 2017
|
|
5K4I
 
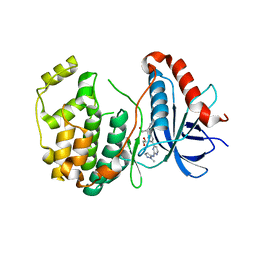 | | Crystal Structure of ERK2 in complex with compound 22 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one, Mitogen-activated protein kinase 1 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-yl)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Inhibitor in Early Clinical Development.
J.Med.Chem., 59, 2016
|
|
9ELF
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron KP.3.1.1 spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Feng, Z, Huang, J, Ward, A.B. | | Deposit date: | 2024-12-04 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural and functional insights into the evolution of SARS-CoV-2 KP.3.1.1 spike protein.
Cell Rep, 44, 2025
|
|
5ECE
 
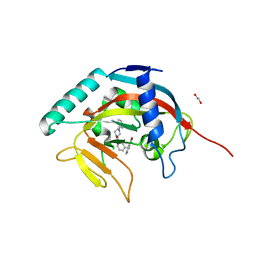 | | Tankyrase 1 with Phthalazinone 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[3-[(4-oxidanylidene-3~{H}-phthalazin-1-yl)methyl]phenyl]carbonylpiperazin-1-yl]pyridine-3-carbonitrile, ACETATE ION, ... | | Authors: | Kazmirski, S.L, Johannes, J, Read, J.A, Howard, T, Larsen, N.A, Ferguson, A.D. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
9ELH
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron KP.3.1.1 spike protein (one RBD up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Feng, Z, Huang, J, Ward, A.B. | | Deposit date: | 2024-12-04 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural and functional insights into the evolution of SARS-CoV-2 KP.3.1.1 spike protein.
Cell Rep, 44, 2025
|
|
5JUG
 
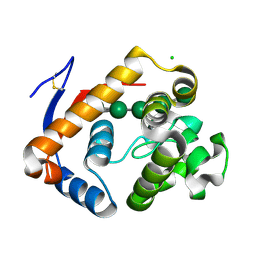 | | Structure of an inactive (E45Q) variant of a beta-1,4-mannanase, SsGH134, in complex with Man5 | | Descriptor: | CHLORIDE ION, GLYCEROL, alpha-D-mannopyranose, ... | | Authors: | Jin, Y, Petricevic, M, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2016-05-10 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | A beta-Mannanase with a Lysozyme-like Fold and a Novel Molecular Catalytic Mechanism.
ACS Cent Sci, 2, 2016
|
|
9ELI
 
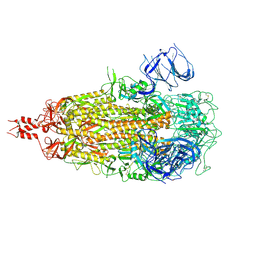 | | Cryo-EM structure of SARS-CoV-2 Omicron KP.3.1.1 spike protein (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Feng, Z, Huang, J, Ward, A.B. | | Deposit date: | 2024-12-04 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural and functional insights into the evolution of SARS-CoV-2 KP.3.1.1 spike protein.
Cell Rep, 44, 2025
|
|
4MQ7
 
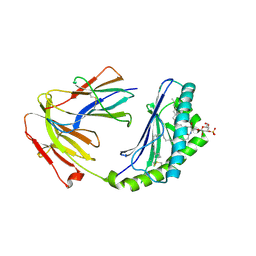 | | Structure of human CD1d-sulfatide | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Luoma, A.M, Adams, E.J. | | Deposit date: | 2013-09-15 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6032 Å) | | Cite: | Crystal Structure of V delta 1 T Cell Receptor in Complex with CD1d-Sulfatide Shows MHC-like Recognition of a Self-Lipid by Human gamma delta T Cells.
Immunity, 39, 2013
|
|
9ELP
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1.11.1+S31 deletion spike protein (one RBD up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Feng, Z, Huang, J, Ward, A.B. | | Deposit date: | 2024-12-04 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural and functional insights into the evolution of SARS-CoV-2 KP.3.1.1 spike protein.
Cell Rep, 44, 2025
|
|
3WPY
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase mutant T47S/S200T/F447V/P500S | | Descriptor: | Beta-fructofuranosidase | | Authors: | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-01-17 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
