9ATG
 
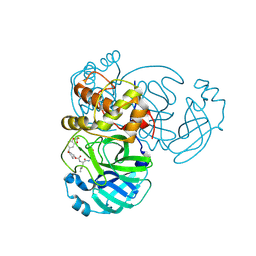 | | Crystal structure of MERS 3CL protease in complex with a 2,2-difluoro-5-methylbenzo[1,3]dioxole 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(2,2-difluoro-2H-1,3-benzodioxol-5-yl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(2,2-difluoro-2H-1,3-benzodioxol-5-yl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
8J52
 
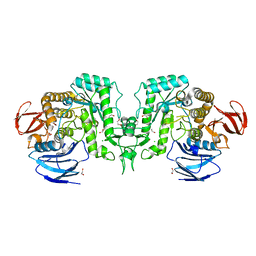 | | Crystal structure of Flavihumibacter petaseus GH31 alpha-galactosidase mutant D304A in complex with alpha-1,4-galactobiose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GH31 alpha-galactosidase, ... | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2023-04-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function analysis of bacterial GH31 alpha-galactosidases specific for alpha-(1→4)-galactobiose.
Febs J., 290, 2023
|
|
5ACB
 
 | | Crystal Structure of the Human Cdk12-Cyclink Complex | | Descriptor: | CYCLIN-DEPENDENT KINASE 12, CYCLIN-K, N-[4-[(3R)-3-[[5-chloranyl-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]piperidin-1-yl]carbonylphenyl]-4-(dimethylamino)butanamide | | Authors: | Dixon Clarke, S.E, Elkins, J.M, Pike, A.C.W, Mackenzie, A, Goubin, S, Strain-Damerell, C, Mahajan, P, Tallant, C, Chalk, R, Wiggers, H, Kopec, J, Fitzpatrick, F, Burgess-Brown, N, Carpenter, E.P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-08-14 | | Release date: | 2016-06-15 | | Last modified: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Covalent Targeting of Remote Cysteine Residues to Develop Cdk12 and Cdk13 Inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
9ATI
 
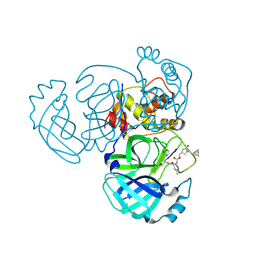 | | Crystal structure of MERS 3CL protease in complex with a racemic bicyclo[2.2.1]heptenyl-methyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-{[N-({[(3S)-1-{[(1R,2R,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(3S)-1-{[(1R,2R,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
5ILH
 
 | | Tobacco 5-epi-aristolochene synthase with CAPSO buffer molecule and Ca2+ ions | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 5-epi-aristolochene synthase, ... | | Authors: | Koo, H.J, Louie, G.V, Xu, Y, Bowman, M, Noel, J.P. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small-molecule buffer components can directly affect terpene-synthase activity by interacting with the substrate-binding site of the enzyme
To Be Published
|
|
8ZHA
 
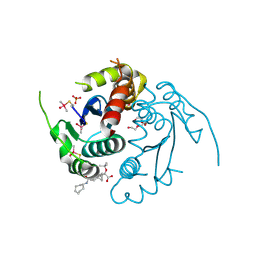 | | HIV-1 integrase core domain in complex with compound 15 | | Descriptor: | (2~{S})-2-[7-(cycloheptylcarbamoyl)-4',5-dimethyl-spiro[1,2-dihydroindene-3,1'-cyclohexane]-4-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, Integrase, SULFATE ION, ... | | Authors: | Furuzono, T, Orita, T, Nomura, A, Adachi, T. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design and synthesis of novel and potent allosteric HIV-1 integrase inhibitors with a spirocyclic moiety.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
8J53
 
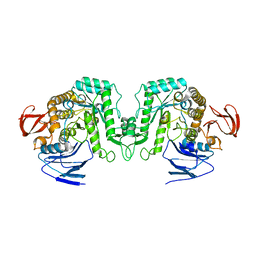 | |
5IIO
 
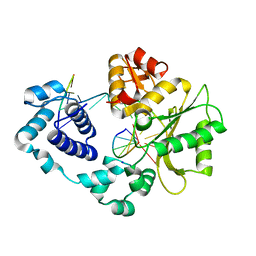 | | Crystal structure of the DNA polymerase lambda binary complex | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*(8OG)P*GP*TP*AP*CP*TP*G)-3'), DNA (5'-D(P*GP*CP*CP*G)-3'), ... | | Authors: | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-17 | | Last modified: | 2016-09-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A fidelity mechanism in DNA polymerase lambda promotes error-free bypass of 8-oxo-dG.
Embo J., 35, 2016
|
|
5A7V
 
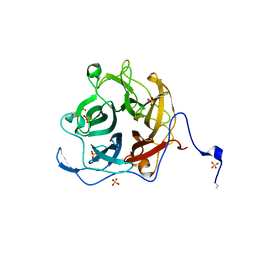 | | The GH130 family of mannoside phosphorylases contains glycoside hydrolases that target beta-1,2 mannosidic linkages in Candida mannan | | Descriptor: | PUTATIVE GLYCOSIDASE PH117-RELATED, SULFATE ION, alpha-D-mannopyranose, ... | | Authors: | Cuskin, F, Basle, A, Day, A.M, Ladeveze, S, Potocki-Veronese, G, Davies, G.J, Gilbert, H.J, Lowe, E. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Gh130 Family of Mannoside Phosphorylases Contains Glycoside Hydrolases that Target Beta-1,2 Mannosidic Linkages in Candida Mannan
J.Biol.Chem., 290, 2015
|
|
9ASW
 
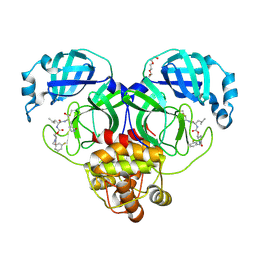 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a m-fluorobenzyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(3-fluorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(3-fluorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ATH
 
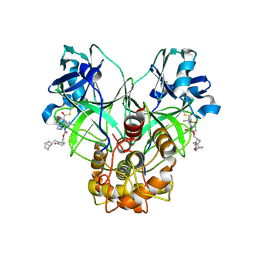 | | Crystal structure of MERS 3CL protease in complex with a methylbicyclo[2.2.1]heptene 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-{[N-({[(2S)-1-{[(1R,2S,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-{[(1R,2S,4R)-bicyclo[2.2.1]hept-5-en-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9BK3
 
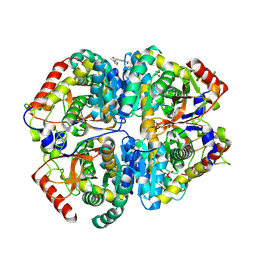 | | Crystal structure of Lactate dehydrogenase in complex with 4-((4-(1-methyl-1H-imidazole-2-carbonyl)phenyl)amino)-4-oxo-2-(4-(trifluoromethyl)phenyl)butanoic acid (R-enantiomer, orthorhombic P form) | | Descriptor: | (2R)-4-[4-(1-methyl-1H-imidazole-2-carbonyl)anilino]-4-oxo-2-[4-(trifluoromethyl)phenyl]butanoic acid, CHLORIDE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Sharma, H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer.
Eur.J.Med.Chem., 275, 2024
|
|
5B5Q
 
 | |
9BNR
 
 | | 4-(2-isothiocyanatoethyl)benzenesulfonamide complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | Macrophage migration inhibitory factor, N-[2-(4-sulfamoylphenyl)ethyl]methanethioamide, SULFATE ION | | Authors: | Fellner, M, Rutledge, M.T, Putha, L, Kok, L.K, Gamble, A.B, Wilbanks, S.M, Vernall, A.J, Tyndall, J.D.A. | | Deposit date: | 2024-05-02 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Covalent isothiocyanate inhibitors of macrophage migration inhibitory factor as potential colorectal cancer treatments.
Chemmedchem, 2024
|
|
5IJR
 
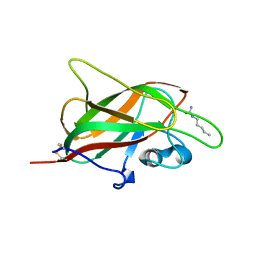 | | X-ray structure of neuropilin-1 b1 domain complexed with Arg-1 ligand. | | Descriptor: | DIMETHYL SULFOXIDE, L-HOMOARGININE, Neuropilin-1 | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-03-02 | | Release date: | 2017-03-29 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
5IK4
 
 | | Laminin A2LG45 C-form, Apo. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Briggs, D.C, Hohenester, E, Campbell, K.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural basis of laminin binding to the LARGE glycans on dystroglycan.
Nat.Chem.Biol., 12, 2016
|
|
9B7C
 
 | |
5IKK
 
 | | Structure of the histone deacetylase Clr3 | | Descriptor: | 1,2-ETHANEDIOL, Histone deacetylase clr3, MAGNESIUM ION, ... | | Authors: | Brugger, C, Schalch, T. | | Deposit date: | 2016-03-03 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SHREC Silences Heterochromatin via Distinct Remodeling and Deacetylation Modules.
Mol.Cell, 62, 2016
|
|
5IQY
 
 | | Structure of apo-Dehydroascorbate Reductase from Pennisetum Glaucum phased by Iodide-SAD method | | Descriptor: | Dehydroascorbate reductase, IODIDE ION | | Authors: | Das, B.K, Kumar, A, Manidola, P, Arockiasamy, A. | | Deposit date: | 2016-03-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Non-native ligands define the active site of Pennisetum glaucum (L.) R. Br dehydroascorbate reductase
Biochem.Biophys.Res.Commun., 473, 2016
|
|
8ZH4
 
 | | HIV-1 integrase core domain in complex with compound 5 | | Descriptor: | (2~{S})-2-(4',5-dimethylspiro[1,2-dihydroindene-3,1'-cyclohexane]-4-yl)-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, Integrase, SULFATE ION, ... | | Authors: | Furuzono, T, Orita, T, Nomura, A, Adachi, T. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design and synthesis of novel and potent allosteric HIV-1 integrase inhibitors with a spirocyclic moiety.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
5I3L
 
 | | DPF3b in complex with H3K14ac peptide | | Descriptor: | 1,2-ETHANEDIOL, H3K14ac peptide, SODIUM ION, ... | | Authors: | Tempel, W, Liu, Y, Walker, J.R, Zhao, A, Qin, S, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-10 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of DPF3b in complex with an acetylated histone peptide.
J.Struct.Biol., 195, 2016
|
|
9B0B
 
 | |
9ASY
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a m-chlorobenzyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(3-chlorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(3-chlorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
5BOQ
 
 | | Human insulin with intra-chain chemical crosslink between modified B24 and B29 | | Descriptor: | Insulin, SULFATE ION | | Authors: | Brzozowski, A.M, Turkenburg, J.P, Jiracek, J, Zakova, L. | | Deposit date: | 2015-05-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational steering of insulin binding specificity by intra-chain chemical crosslinking.
Sci Rep, 6, 2016
|
|
9ATE
 
 | | Crystal structure of MERS 3CL protease in complex with a methylbicyclo[2.2.1]heptane 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-{[N-({[(2S)-1-{[(1S,2S,4R)-bicyclo[2.2.1]heptan-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-{[(1S,2S,4R)-bicyclo[2.2.1]heptan-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
