4WTR
 
 | | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaribiose | | Descriptor: | beta-1,3-glucanosyltransferase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Qin, Z, Yan, Q, Lei, J, Yang, S, Jiang, Z. | | Deposit date: | 2014-10-30 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The first crystal structure of a glycoside hydrolase family 17 beta-1,3-glucanosyltransferase displays a unique catalytic cleft.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8HOO
 
 | |
4WJ8
 
 | | Human Pyruvate Kinase M2 Mutant C424A | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Mitchell, T, Yuan, M, McNae, I, Morgan, H, Walkinshaw, M.D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Human Pyruvate Kinase M2 Mutant C424A
To Be Published
|
|
8HPY
 
 | | Crystal structure of human LGI1-ADAM22 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Disintegrin and metalloproteinase domain-containing protein 22, ... | | Authors: | Liu, H, Xu, F. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (5.87 Å) | | Cite: | Crystal structure of human LGI1-ADAM22 complex
To Be Published
|
|
4WJS
 
 | |
8HFV
 
 | | Crystal structure of CTSL in complex with K777 | | Descriptor: | CACODYLATE ION, Nalpha-[(4-methylpiperazin-1-yl)carbonyl]-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-phenylalaninamide, Procathepsin L, ... | | Authors: | Wang, H, Shao, M, Sun, L, Yang, H. | | Deposit date: | 2022-11-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
4WK0
 
 | | Metal Ion and Ligand Binding of Integrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARG-GLY-ASP, ... | | Authors: | Xia, W, Springer, T.A. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Metal ion and ligand binding of integrin alpha 5 beta 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8HMT
 
 | | The complex of ACK1 with the inhibitor 2-142 | | Descriptor: | 6-(2-bromophenyl)-2-[[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-8-[[(2S)-oxolan-2-yl]methyl]pyrido[2,3-d]pyrimidin-7-one, Activated CDC42 kinase 1 | | Authors: | Zhu, S, Xiaoyun, X.Y. | | Deposit date: | 2022-12-05 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | The complex of ACK1 with the inhibitor 2-142
To Be Published
|
|
4WTU
 
 | | Crystal structure of BACE1 in complex with 2-aminooxazoline 3-aza-4-fluoro-xanthene inhibitor 22 | | Descriptor: | (5S)-3-(5,6-dihydro-2H-pyran-3-yl)-1-fluoro-7-(2-fluoropyridin-3-yl)spiro[chromeno[2,3-c]pyridine-5,4'-[1,3]oxazol]-2'-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2014-10-30 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Orally Available BACE1 Inhibitor That Affords Robust CNS A beta Reduction without Cardiovascular Liabilities.
Acs Med.Chem.Lett., 6, 2015
|
|
8HOT
 
 | |
8HLT
 
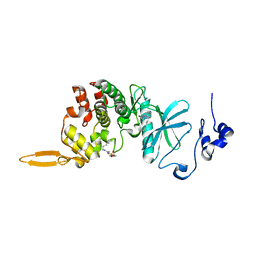 | | The co-crystal structure of DYRK2 with YK-2-99B | | Descriptor: | (6-{[(4P)-4-(1,3-benzothiazol-5-yl)-5-fluoropyrimidin-2-yl]amino}pyridin-3-yl)(piperazin-1-yl)methanone, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Shen, H.T, Xiao, Y.B, Yuan, K, Yang, P, Li, Q.N. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Potent DYRK2 Inhibitors with High Selectivity, Great Solubility, and Excellent Safety Properties for the Treatment of Prostate Cancer.
J.Med.Chem., 66, 2023
|
|
4WTY
 
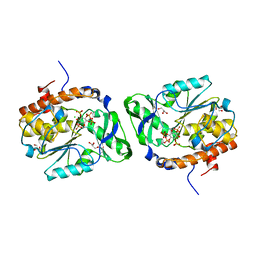 | | Structure of the PTP-like myo-inositol phosphatase from Selenomonas ruminantium in complex with myo-inositol-(1,3,4,5)-tetrakisphosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, ... | | Authors: | Bruder, L.M, Mosimann, S.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol-(1,3,4,5)-tetrakisphosphate
To Be Published
|
|
4WU3
 
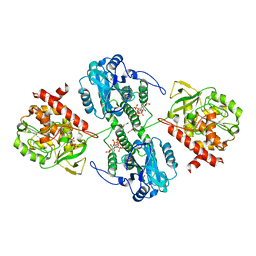 | | Structure of the PTP-like myo-inositol phosphatase from Mitsuokella multacida in complex with myo-inositol-(1,3,4,5)-tetrakisphosphate | | Descriptor: | GLYCEROL, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, MYO-INOSITOL PHOSPHOHYDROLASE, ... | | Authors: | Bruder, L.M, Mosimann, S.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol-(1,3,4,5)-tetrakisphosphate
To Be Published
|
|
8HOP
 
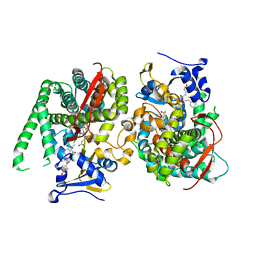 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Nap-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Nap-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CW4
 
 | | CryoEM structure of the N-pilus from Escherichia coli | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Conjugal transfer protein TraM | | Authors: | Bui, K.H, Black, C.S. | | Deposit date: | 2022-05-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of the Agrobacterium tumefaciens T-pilus reveals the importance of positive charges in the lumen.
Structure, 31, 2023
|
|
8HOQ
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Phe(4CF3)-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Phe(4CF3)-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4WKO
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with hydroxybutylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-hydroxybutyl)sulfanyl]methyl}pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase | | Authors: | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
4WEQ
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc02828 (SmGhrA) from Sinorhizobium meliloti in complex with NADP and sulfate | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sroka, P, Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Osinski, T, Hillerich, B.S, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-10 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
8HQ1
 
 | |
4WF5
 
 | | Crystal structure of E.Coli DsbA soaked with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazole-5-carboxylic acid, COPPER (II) ION, ... | | Authors: | Adams, L.A, Sharma, P, Mohanty, B, Ilyichova, O.V, Mulcair, M.D, Williams, M.L, Gleeson, E.C, Totsika, M, Doak, B.C, Caria, S, Rimmer, K, Shouldice, S.R, Vazirani, M, Headey, S.J, Plumb, B.R, Martin, J.L, Heras, B, Simpson, J.S, Scanlon, M.J. | | Deposit date: | 2014-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8HP3
 
 | | Crystal structure of meso-diaminopimelate dehydrogenase from Prevotella timonensis | | Descriptor: | 1,2-ETHANEDIOL, Meso-diaminopimelate D-dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tan, Y, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Rational Design of Meso -Diaminopimelate Dehydrogenase with Enhanced Reductive Amination Activity for Efficient Production of d- p -Hydroxyphenylglycine.
Appl.Environ.Microbiol., 89, 2023
|
|
8HON
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with Im-C6-Tyr-Tyr | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-C6-Tyr-Tyr, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4WV9
 
 | | Crystal structure of acetylcholine binding protein (AChBP) from Aplysia Californica in complex with click chemistry compound (3-exo)-8,8-dimethyl-3-[4-(pyridin-4-yl)-1H-1,2,3-triazol-1-yl]-8-azoniabicyclo[3.2.1]octane | | Descriptor: | (3-exo)-8,8-dimethyl-3-[4-(pyridin-4-yl)-1H-1,2,3-triazol-1-yl]-8-azoniabicyclo[3.2.1]octane, Soluble acetylcholine receptor | | Authors: | Talley, T.T, Bobango, J, Wu, J.M, Sankaran, B. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of acetylcholine binding protein (AChBP) from Aplysia Californica in complex with click chemistry compound.
To Be Published
|
|
8HP0
 
 | | Crystal structure of meso-diaminopimelate dehydrogenase from Prevotella timonensis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Meso-diaminopimelate D-dehydrogenase, SULFATE ION | | Authors: | Tan, Y, Song, W. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Rational Design of Meso -Diaminopimelate Dehydrogenase with Enhanced Reductive Amination Activity for Efficient Production of d- p -Hydroxyphenylglycine.
Appl.Environ.Microbiol., 89, 2023
|
|
4WVR
 
 | | Crystal structure of Dscam1 Ig7 domain, isoform 5 | | Descriptor: | Down syndrome cell adhesion molecule, isoform AK | | Authors: | Chen, Q, Yu, Y, Li, S, Cheng, L. | | Deposit date: | 2014-11-07 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
