3IDI
 
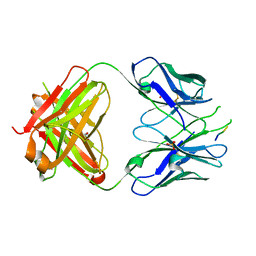 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 Fab' fragment in complex with gp41 Peptide ALDKWNQ | | Descriptor: | 2F5 Fab heavy chain, 2F5 Fab light chain, GLYCEROL, ... | | Authors: | Julien, J.-P, Bryson, S, Pai, E.F. | | Deposit date: | 2009-07-21 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications.
J.Virol., 83, 2009
|
|
4YJK
 
 | | Crystal structure of C212S mutant of Shewanella oneidensis MR-1 uridine phosphorylase | | Descriptor: | SULFATE ION, URACIL, Uridine phosphorylase | | Authors: | Safonova, T.N, Mordkovich, N.N, Manuvera, V.A, Dorovatovsky, P.V, Veiko, V.P, Popov, V.O, Polyakov, K.M. | | Deposit date: | 2015-03-03 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Concerted action of two subunits of the functional dimer of Shewanella oneidensis MR-1 uridine phosphorylase derived from a comparison of the C212S mutant and the wild-type enzyme.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6AXY
 
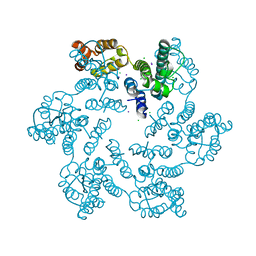 | |
1RZ7
 
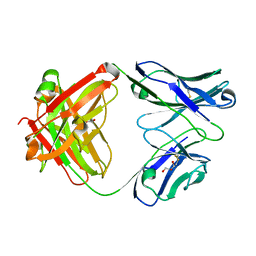 | | CRYSTAL STRUCTURE OF HUMAN ANTI-HIV-1 GP120-REACTIVE ANTIBODY 48D | | Descriptor: | Fab 48d heavy chain, Fab 48d light chain, GLYCEROL | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6FIL
 
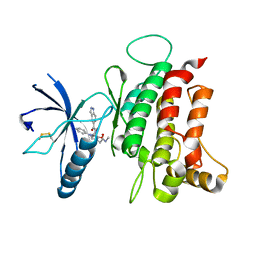 | | DDR1, 2-[8-(1H-indazole-5-carbonyl)-4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-N-methylacetamide, 1.730A, P212121, Rfree=24.5% | | Descriptor: | 2-[8-(2~{H}-indazol-5-ylcarbonyl)-4-oxidanylidene-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-~{N}-methyl-ethanamide, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
1K3O
 
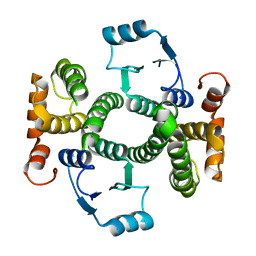 | | Crystal Structure Analysis of apo Glutathione S-Transferase | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1 | | Authors: | Le Trong, I, Stenkamp, R.E, Ibarra, C, Atkins, W.M, Adman, E.T. | | Deposit date: | 2001-10-03 | | Release date: | 2002-10-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.3-A resolution structure of human glutathione S-transferase with S-hexyl glutathione bound reveals possible extended ligandin binding site.
Proteins, 48, 2002
|
|
2VN8
 
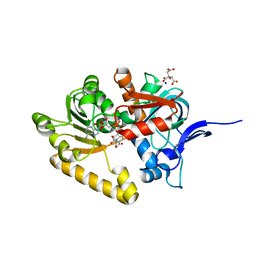 | | Crystal structure of human Reticulon 4 interacting protein 1 in complex with NADPH | | Descriptor: | CITRIC ACID, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pike, A.C.W, Guo, K, Elkins, J, Ugochukwu, E, Roos, A.K, Filippakopoulos, P, von Delft, F, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Oppermann, U. | | Deposit date: | 2008-01-31 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Reticulon 4 Interacting Protein 1 in Complex with Nadph
To be Published
|
|
6UAW
 
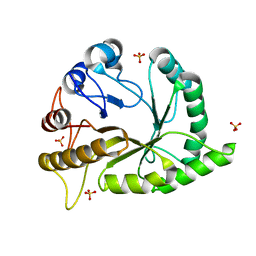 | | Crystal structure of a GH128 (subgroup II) endo-beta-1,3-glucanase from Pseudomonas viridiflava (PvGH128_II) in complex with laminaritriose | | Descriptor: | Glyco_hydro_cc domain-containing protein, SULFATE ION, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Santos, C.R, Costa, P.A.C.R, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
1M0L
 
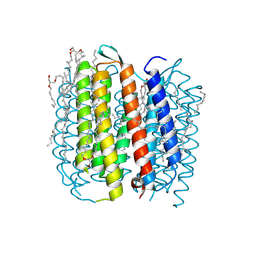 | | BACTERIORHODOPSIN/LIPID COMPLEX AT 1.47 A RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN, ... | | Authors: | Lanyi, J.K. | | Deposit date: | 2002-06-13 | | Release date: | 2002-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic structure of the K intermediate of bacteriorhodopsin: conservation of free energy after photoisomerization of the retinal.
J.Mol.Biol., 321, 2002
|
|
7V19
 
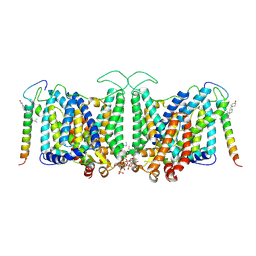 | | Local refinement of Band 3-II transmembrane domains, class 1 of erythrocyte ankyrin-1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CHOLESTEROL, ... | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1M22
 
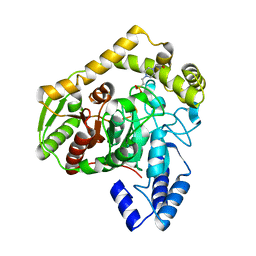 | | X-ray structure of native peptide amidase from Stenotrophomonas maltophilia at 1.4 A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, peptide amidase | | Authors: | Labahn, J, Neumann, S, Buldt, G, Kula, M.-R, Granzin, J. | | Deposit date: | 2002-06-21 | | Release date: | 2002-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An alternative mechanism for amidase signature enzymes
J.MOL.BIOL., 322, 2002
|
|
1RZF
 
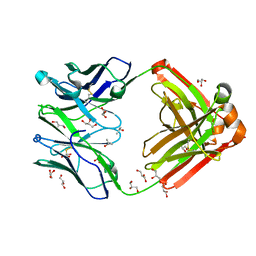 | | Crystal structure of Human anti-HIV-1 GP120-reactive antibody E51 | | Descriptor: | Fab E51 heavy chain, Fab E51 light chain, GLYCEROL, ... | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5EM6
 
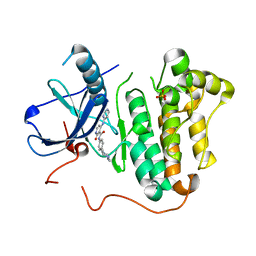 | |
5UZS
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P200 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-{[(4-chlorophenyl)carbamoyl]amino}propan-2-yl)-N-hydroxybenzene-1-carboximidamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.367 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P200
To Be Published
|
|
6AXS
 
 | |
6AY5
 
 | | CREBBP bromodomain in complex with Cpd17 (5-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-3-methylbenzo[d]thiazol-2(3H)-one) | | Descriptor: | 5-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-3-methyl-1,3-benzothiazol-2(3H)-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A Unique Approach to Design Potent and Selective Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP) Inhibitors.
J. Med. Chem., 60, 2017
|
|
2VLB
 
 | | Structure of unliganded arylmalonate decarboxylase | | Descriptor: | 1,2-ETHANEDIOL, ARYLMALONATE DECARBOXYLASE, BETA-MERCAPTOETHANOL, ... | | Authors: | Kuettner, E.B, Keim, A, Kircher, M, Rosmus, S, Strater, N. | | Deposit date: | 2008-01-11 | | Release date: | 2008-03-18 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Active Site Mobility Revealed by the Crystal Structure of Arylmalonate Decarboxylase from Bordetella Bronchiseptica
J.Mol.Biol., 377, 2008
|
|
5EQJ
 
 | | Crystal structure of the two-subunit tRNA m1A58 methyltransferase from Saccharomyces cerevisiae | | Descriptor: | tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRM61, tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 | | Authors: | Zhu, Y, Wang, M, Wang, C, Fan, X, Jiang, X, Teng, M, Li, X. | | Deposit date: | 2015-11-13 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the two-subunit tRNA m(1)A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
5K0N
 
 | | Crystal Structure of COMT in complex with 4-[5-[1-(4-methoxyphenyl)cyclopropyl]-1H-pyrazol-3-yl]-1,3-dimethylpyrazole | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 5-[1-(4-methoxyphenyl)cyclopropyl]-1',3'-dimethyl-1'H,2H-3,4'-bipyrazole, ... | | Authors: | Ehler, A, Rodriguez-Sarmiento, R.M. | | Deposit date: | 2016-05-17 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Design of Potent and Druglike Nonphenolic Inhibitors for Catechol O-Methyltransferase Derived from a Fragment Screening Approach Targeting the S-Adenosyl-l-methionine Pocket.
J. Med. Chem., 59, 2016
|
|
4N71
 
 | | X-Ray Crystal Structure of 2-amino-1-hydroxyethylphosphonate-bound PhnZ | | Descriptor: | FE (III) ION, Predicted HD phosphohydrolase PhnZ, [(1R)-2-amino-1-hydroxyethyl]phosphonic acid | | Authors: | Woersdoerfer, B, Lingaraju, M, Yennawar, N, Boal, A.K, Krebs, C, Bollinger Jr, J.M, Pandelia, M.-E. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | Organophosphonate-degrading PhnZ reveals an emerging family of HD domain mixed-valent diiron oxygenases.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5CVJ
 
 | | Monolignol 4-O-methyltransferase 5 - coniferyl alcohol | | Descriptor: | (Iso)eugenol O-methyltransferase, 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2-methoxyphenol, NITRATE ION, ... | | Authors: | Cai, Y, Liu, C.-J. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of coniferyl alcohol bound monolignol 4-O-methyltransferase at 1.68 Angstroms resolution
To Be Published
|
|
5B0P
 
 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - glycerol complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
4QGD
 
 | | Crystal Structure of the Complex of Phospholipase A2 with Gramine derivative at 1.80 A Resolution | | Descriptor: | 3-{3-[(DIMETHYLAMINO)METHYL]-1H-INDOL-7-YL}PROPAN-1-OL, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Shukla, P.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-22 | | Release date: | 2014-06-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures and binding studies of the complexes of phospholipase A2 with five inhibitors
Biochim.Biophys.Acta, 1854, 2015
|
|
3H1V
 
 | | Human glucokinase in complex with a synthetic activator | | Descriptor: | 1-({5-[4-(methylsulfonyl)phenoxy]-2-pyridin-2-yl-1H-benzimidazol-6-yl}methyl)pyrrolidine-2,5-dione, Glucokinase, SODIUM ION, ... | | Authors: | Kamata, K, Takahashi, K. | | Deposit date: | 2009-04-14 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The design and optimization of a series of 2-(pyridin-2-yl)-1H-benzimidazole compounds as allosteric glucokinase activators.
Bioorg.Med.Chem., 17, 2009
|
|
3JQ0
 
 | |
