9F0F
 
 | |
9EZ8
 
 | | Cryo-EM structure of the icosahedral lumazine synthase from Vicia faba. | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Chee, M, Trapani, S, Hoh, F, Lai Kee Him, J, Yvon, M, Blanc, S, Bron, P. | | Deposit date: | 2024-04-11 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of the icosahedral lumazine synthase from Vicia faba.
To Be Published
|
|
9EY8
 
 | | Crystal structure of human tyrosinase-related protein 1 (TYRP1) in complex with (s)-amino-L-tyrosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ng, Y.M, Soler-Lopez, M. | | Deposit date: | 2024-04-09 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions of phenylalanine derivatives with human tyrosinase: lessons from experimental and theoretical studies.
Chembiochem, 2024
|
|
9EY7
 
 | |
9EY6
 
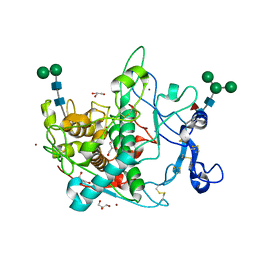 | | Crystal structure of human tyrosinase-related protein 1 (TYRP1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 5,6-dihydroxyindole-2-carboxylic acid oxidase, ... | | Authors: | Ng, Y.M, Soler-Lopez, M. | | Deposit date: | 2024-04-09 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.228 Å) | | Cite: | Interactions of phenylalanine derivatives with human tyrosinase: lessons from experimental and theoretical studies.
Chembiochem, 2024
|
|
9EY5
 
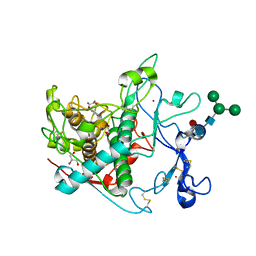 | |
9EXY
 
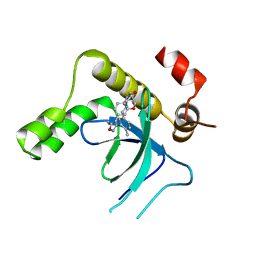 | | Crystal structure of the PWWP1 domain of NSD2 bound by compound 34. | | Descriptor: | 7-[5-methyl-3-[2-methyl-5-(piperidin-1-ylmethyl)phenyl]-1,2-oxazol-4-yl]-4~{H}-1,4-benzoxazin-3-one, Histone-lysine N-methyltransferase NSD2 | | Authors: | Collie, G.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Identification of Novel Potent NSD2-PWWP1 Ligands Using Structure-Based Design and Computational Approaches.
J.Med.Chem., 2024
|
|
9EXX
 
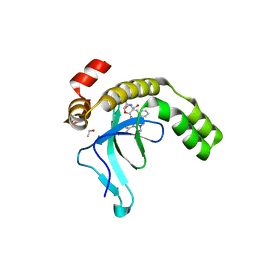 | | Crystal structure of the PWWP1 domain of NSD2 bound by compound 18. | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[1-methyl-5-(3-oxidanylidene-4~{H}-1,4-benzoxazin-7-yl)imidazol-4-yl]-~{N}-phenyl-benzamide, ETHANOL, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Identification of Novel Potent NSD2-PWWP1 Ligands Using Structure-Based Design and Computational Approaches.
J.Med.Chem., 2024
|
|
9EXW
 
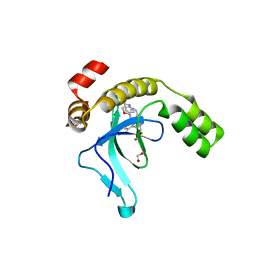 | | Crystal structure of the PWWP1 domain of NSD2 bound by compound 17. | | Descriptor: | 1,2-ETHANEDIOL, 7-[3-methyl-5-[2-methyl-5-[(pyridin-3-ylamino)methyl]phenyl]imidazol-4-yl]-4~{H}-1,4-benzoxazin-3-one, Histone-lysine N-methyltransferase NSD2 | | Authors: | Collie, G.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Identification of Novel Potent NSD2-PWWP1 Ligands Using Structure-Based Design and Computational Approaches.
J.Med.Chem., 2024
|
|
9EWX
 
 | | Cryo-EM structure of the Pseudomonas aeruginosa PAO1 Type IV pilus | | Descriptor: | Pilin | | Authors: | Ochner, H, Boehning, J, Wang, Z, Tarafder, A, Caspy, I, Bharat, T.A.M. | | Deposit date: | 2024-04-05 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structure of the Pseudomonas aeruginosa PAO1 Type-IV pilus
To Be Published
|
|
9EWQ
 
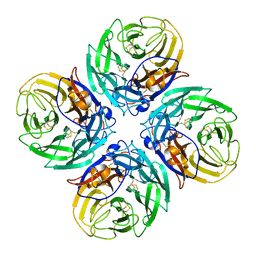 | |
9EWO
 
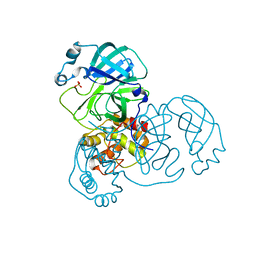 | | Mpro from SARS-CoV-2 with R4A R298A double mutations | | Descriptor: | Non-structural protein 11, SULFATE ION | | Authors: | Plewka, J, Lis, K, Chykunova, Y, Czarna, A, Kantyka, T, Pyrc, K. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
9EWN
 
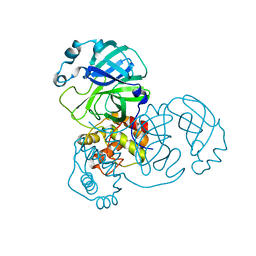 | | Mpro from SARS-CoV-2 with 4Q mutation | | Descriptor: | Non-structural protein 11 | | Authors: | Plewka, J, Lis, K, Czarna, A, Kantyka, T, Pyrc, K. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
9EWM
 
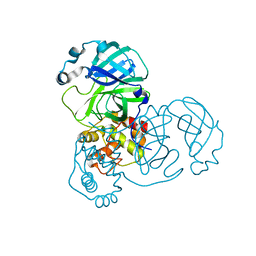 | | Mpro from SARS-CoV-2 with R4Q R298Q double mutations | | Descriptor: | Non-structural protein 11 | | Authors: | Plewka, J, Lis, K, Chykunova, Y, Czarna, A, Kantyka, T, Pyrc, K. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
9EUS
 
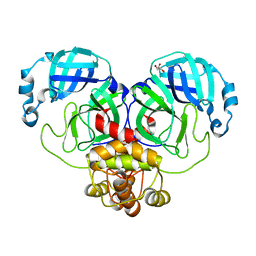 | | Mpro from SARS-CoV-2 with R298A mutation | | Descriptor: | GLYCEROL, Replicase polyprotein 1a | | Authors: | Plewka, J, Lis, K, Czarna, A, Pyrc, K, Kantyka, T, Chykunova, Y. | | Deposit date: | 2024-03-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
9EUR
 
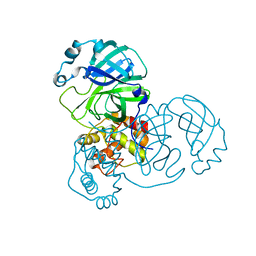 | | Mpro WT from SARS-CoV-2 with 298Q mutation | | Descriptor: | Replicase polyprotein 1a | | Authors: | Plewka, J, Lis, K, Czarna, A, Pyrc, K, Kantyka, T, Chykunova, Y. | | Deposit date: | 2024-03-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
9EUN
 
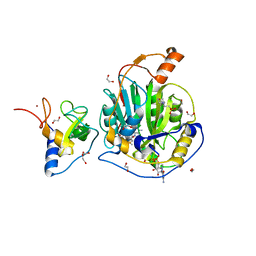 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with SAM and m7GTP | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
9EST
 
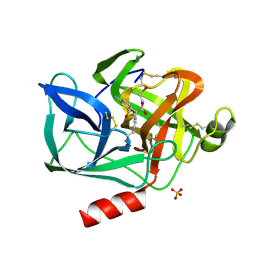 | | STRUCTURAL STUDY OF PORCINE PANCREATIC ELASTASE COMPLEXED WITH 7-AMINO-3-(2-BROMOETHOXY)-4-CHLOROISOCOUMARIN AS A NONREACTIVATABLE DOUBLY COVALENT ENZYME-INHIBITOR COMPLEX | | Descriptor: | (2-BROMOETHYL)(2-'FORMYL-4'-AMINOPHENYL) ACETATE, CALCIUM ION, PORCINE PANCREATIC ELASTASE, ... | | Authors: | Radhakrishnan, R, Powers, J.C, Meyer Jr, E.F. | | Deposit date: | 1991-01-14 | | Release date: | 1994-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural study of porcine pancreatic elastase complexed with 7-amino-3-(2-bromoethoxy)-4-chloroisocoumarin as a nonreactivatable doubly covalent enzyme-inhibitor complex.
Biochemistry, 30, 1991
|
|
9ERO
 
 | |
9ERN
 
 | |
9ERM
 
 | |
9ER3
 
 | |
9EQK
 
 | |
9EQH
 
 | | WWP2 WW2-2,3-linker-HECT (WWP2-LH) | | Descriptor: | GLYCEROL, Isoform 2 of NEDD4-like E3 ubiquitin-protein ligase WWP2, SODIUM ION | | Authors: | Dudey, A.P, Hemmings, A.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Expanding the Inhibitor Space of the WWP1 and WWP2 HECT E3 Ligases
To Be Published
|
|
9EQ4
 
 | | Structure of IgE HMM5 bound to FceRIa cryo-EM class 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, ... | | Authors: | Andersen, G.R, Jensen, R.K. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structure of IgE HMM5 bound to FceRIa cryo-EM class 5
To be published
|
|
