1M5W
 
 | | 1.96 A Crystal Structure of Pyridoxine 5'-Phosphate Synthase in Complex with 1-deoxy-D-xylulose phosphate | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, PHOSPHATE ION, Pyridoxal phosphate biosynthetic protein pdxJ | | Authors: | Yeh, J.I, Du, S, Pohl, E, Cane, D.E. | | Deposit date: | 2002-07-10 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Multistate Binding in Pyridoxine 5'-Phosphate Synthase: 1.96 A Crystal Structure in
Complex with 1-deoxy-D-xylulose phosphate
Biochemistry, 41, 2002
|
|
4AVS
 
 | |
4CSC
 
 | |
4MIC
 
 | | Crystal structure of Gpb in complex with SUGAR (N-{(2E)-3-[4-(PROPAN-2-YL)PHENYL]PROP-2-ENOYL}-BETA-D-GLUCOPYRANOSYLAMINE) (S6) | | Descriptor: | Glycogen phosphorylase, muscle form, N-{(2E)-3-[4-(propan-2-yl)phenyl]prop-2-enoyl}-beta-D-glucopyranosylamine | | Authors: | Kantsadi, A.L, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
4MI6
 
 | | Crystal structure of Gpb in complex with SUGAR (N-[4-(5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)BUTANOYL]-BETA-D-GLUCOPYRANOSYLAMINE) | | Descriptor: | Glycogen phosphorylase, muscle form, N-[4-(5,6,7,8-tetrahydronaphthalen-2-yl)butanoyl]-beta-D-glucopyranosylamine | | Authors: | Kantsadi, L.A, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
4MHS
 
 | | Crystal structure of Gpb in complex with SUGAR (N-[(2E)-3-(BIPHENYL-4-YL)PROP-2-ENOYL]-BETA-D-GLUCOPYRANOSYLAMINE | | Descriptor: | DIMETHYL SULFOXIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Kantsadi, L.A, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
4MI3
 
 | | Crystal structure of Gpb in complex with SUGAR (N-{(2R)-2-METHYL-3-[4-(PROPAN-2-YL)PHENYL]PROPANOYL}-BETA-D-GLUCOPYRANOSYLAMINE) (S21) | | Descriptor: | Glycogen phosphorylase, muscle form, N-{(2R)-2-methyl-3-[4-(propan-2-yl)phenyl]propanoyl}-beta-D-glucopyranosylamine | | Authors: | Kantsadi, L.A, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
4MHO
 
 | | Crystal structure of Gpb in complex with S3, SUGAR (N-[(BIPHENYL-4-YLOXY)ACETYL]-BETA-D-GLUCOPYRANOSYLAMINE) | | Descriptor: | Glycogen phosphorylase, muscle form, N-[(biphenyl-4-yloxy)acetyl]-beta-D-glucopyranosylamine | | Authors: | Kantsadi, A.L, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
4MI9
 
 | | Crystal structure of Gpb in complex with SUGAR (N-[(3R)-3-(4-ETHYLPHENYL)BUTANOYL]-BETA-D-GLUCOPYRANOSYLAMINE) (S20) | | Descriptor: | Glycogen phosphorylase, muscle form, N-[(3R)-3-(4-ethylphenyl)butanoyl]-beta-D-glucopyranosylamine | | Authors: | Kantsadi, A.L, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
3QKE
 
 | | Crystal structure of D-mannonate dehydratase from Chromohalobacter Salexigens complexed with Mg and D-Gluconate | | Descriptor: | D-gluconic acid, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-02-01 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cystal structure of D-mannonate dehydratase from Chromohalobacter salexigens complexed with Mg and D-Gluconate
To be Published
|
|
1XLG
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | ALUMINUM ION, D-XYLOSE ISOMERASE, MAGNESIUM ION, ... | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XIE
 
 | |
4LED
 
 | | The Crystal Structure of Pyocin L1 bound to D-rhamnose at 2.37 Angstroms | | Descriptor: | Pyocin L1, alpha-D-rhamnopyranose | | Authors: | Grinter, R, Roszak, A.W, Mccaughey, L, Cogdell, C.J, Walker, D. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Lectin-Like Bacteriocins from Pseudomonas spp. Utilise D-Rhamnose Containing Lipopolysaccharide as a Cellular Receptor.
Plos Pathog., 10, 2014
|
|
4Q6Q
 
 | | Structural analysis of the Zn-form II of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2UUO
 
 | | Crystal structure of MurD ligase in complex with D-Glu containing sulfonamide inhibitor | | Descriptor: | N-{[6-(PENTYLOXY)NAPHTHALEN-2-YL]SULFONYL}-D-GLUTAMIC ACID, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Humljan, J, Kotnik, M, Contreras-Martel, C, Blanot, D, Urleb, U, Dessen, A, Solmajer, T, Gobec, S. | | Deposit date: | 2007-03-06 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel naphthalene-N-sulfonyl-D-glutamic acid derivatives as inhibitors of MurD, a key peptidoglycan biosynthesis enzyme.
J. Med. Chem., 51, 2008
|
|
2UUP
 
 | | Crystal structure of MurD ligase in complex with D-Glu containing sulfonamide inhibitor | | Descriptor: | N-({6-[(4-CYANOBENZYL)OXY]NAPHTHALEN-2-YL}SULFONYL)-D-GLUTAMIC ACID, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Humljan, J, Kotnik, M, Contreras-Martel, C, Blanot, D, Urleb, U, Dessen, A, Solmajer, T, Gobec, S. | | Deposit date: | 2007-03-06 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Novel naphthalene-N-sulfonyl-D-glutamic acid derivatives as inhibitors of MurD, a key peptidoglycan biosynthesis enzyme.
J. Med. Chem., 51, 2008
|
|
2GMU
 
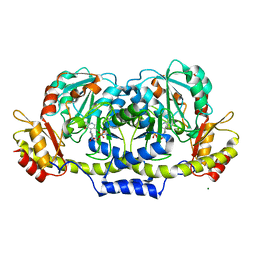 | | Crystal structure of E coli GDP-4-keto-6-deoxy-D-mannose-3-dehydratase complexed with PLP-glutamate ketimine intermediate | | Descriptor: | MAGNESIUM ION, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-D-GLUTAMIC ACID, Putative pyridoxamine 5-phosphate-dependent dehydrase, ... | | Authors: | Cook, P.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2006-04-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of GDP-4-keto-6-deoxy-D-mannose-3-dehydratase: a unique coenzyme B6-dependent enzyme.
Protein Sci., 15, 2006
|
|
4Q6M
 
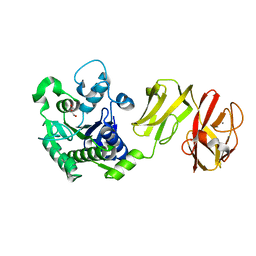 | | Structural analysis of the apo-form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | CALCIUM ION, Conserved hypothetical secreted protein, GLYCEROL | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6O
 
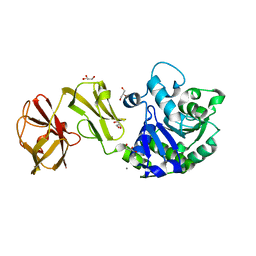 | | Structural analysis of the mDAP-bound form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q6P
 
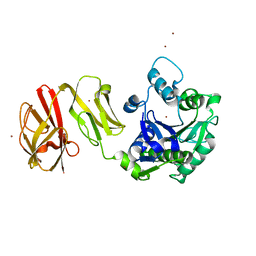 | | Structural analysis of the Zn-form I of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, CALCIUM ION, Conserved hypothetical secreted protein, ... | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3BD6
 
 | | Glycogen Phosphorylase complex with 1(-D-ribofuranosyl) cyanuric acid | | Descriptor: | 1-beta-D-ribofuranosyl-1,3,5-triazinane-2,4,6-trione, Glycogen phosphorylase, muscle form | | Authors: | Sovantzis, D.A, Hadjiloi, T, Hayes, J.M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D-Glucopyranosyl pyrimidine nucleoside binding to muscle glycogen phosphorylase b
To be Published
|
|
2W92
 
 | | Structure of a Streptococcus pneumoniae family 85 glycoside hydrolase, Endo-D, in complex with NAG-thiazoline. | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ENDO-BETA-N-ACETYLGLUCOSAMINIDASE D, PENTAETHYLENE GLYCOL | | Authors: | Abbott, D.W, Macauley, M.S, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2009-01-21 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Streptococcus Pneumoniae Endohexosaminidase D, Structural and Mechanistic Insight Into Substrate-Assisted Catalysis in Family 85 Glycoside Hydrolases.
J.Biol.Chem., 284, 2009
|
|
1CZQ
 
 | | CRYSTAL STRUCTURE OF THE D10-P1/IQN17 COMPLEX: A D-PEPTIDE INHIBITOR OF HIV-1 ENTRY BOUND TO THE GP41 COILED-COIL POCKET. | | Descriptor: | CHLORIDE ION, D-PEPTIDE INHIBITOR, FUSION PROTEIN BETWEEN THE HYDROPHOBIC POCKET OF HIV GP41 AND GCN4-PIQI | | Authors: | Eckert, D.M, Malashkevich, V.N, Hong, L.H, Carr, P.A, Kim, P.S. | | Deposit date: | 1999-09-06 | | Release date: | 1999-10-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket.
Cell(Cambridge,Mass.), 99, 1999
|
|
3S0J
 
 | | The crystal structure of glycogen phosphorylase b in complex with 2,5-dihydroxy-4-(beta-D-glucopyranosyl)-chlorobenzene | | Descriptor: | (1S)-1,5-anhydro-1-(4-chloro-2,5-dihydroxyphenyl)-D-glucitol, Glycogen phosphorylase, muscle form | | Authors: | Alexacou, K.-M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G, Leonidas, D.D. | | Deposit date: | 2011-05-13 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Halogen-substituted (C-beta-D-glucopyranosyl)-hydroquinone regioisomers: synthesis, enzymatic evaluation and their binding to glycogen phosphorylase.
Bioorg.Med.Chem., 19, 2011
|
|
6LL9
 
 | |
